2. Package Documentation¶
The modlamp package includes the following modules:
Module |
Characteristics |
|---|---|
Module incorporating |
|
Module incorporating different classes for generating peptide sequences with different characteristics. |
|
Module incorporating functions to connect to the modlab in-house peptide database server. |
|
Module incorporating functions to load different peptide datasets used for ML classification tasks. |
|
Module incorporating functions to visualize descriptor. |
|
Module incorporating machine learning functions for training SVM or RF models on peptides. |
|
Module incorporating classes for analysis of wetlab data like circular dichroism plots. |
|
Module incorporating the |
|
Module incorporating helper functions for other modules. |
2.1. modlamp.descriptors¶
This module incorporates different classes to calculate peptide descriptor values. The following classes are available:
Class |
Characteristics |
|---|---|
Global one-dimensional peptide descriptors calculated from the AA sequence. |
|
AA scale based global or convoluted descriptors (auto-/cross-correlated). |
See also
modlamp.core.BaseDescriptor from which the classes in modlamp.descriptors inherit.
-
class
modlamp.descriptors.GlobalDescriptor(seqs)[source]¶ Base class for global, non-amino acid scale dependant descriptors. The following descriptors can be calculated by the methods linked below:
-
length(append=False)[source]¶ Method to calculate the length (total AA count) of every sequence in the attribute
sequences.- Parameters
append – {boolean} whether the produced descriptor values should be appended to the existing ones in the attribute
descriptor.- Returns
array of sequence lengths in the attribute
descriptor- Example
>>> desc = GlobalDescriptor(['AFDGHLKI','KKLQRSDLLRTK','KKLASCNNIPPR']) >>> desc.length() >>> desc.descriptor array([[ 8.], [12.], [12.]])
-
formula(amide=False, append=False)[source]¶ Method to calculate the molecular formula of every sequence in the attribute
sequences.- Parameters
amide – {boolean} whether the sequences are C-terminally amidated.
append – {boolean} whether the produced descriptor values should be appended to the existing ones in the attribute
descriptor.
- Returns
array of molecular formulas {str} in the attribute
descriptor- Example
>>> desc = GlobalDescriptor(['KADSFLSADGHSADFSLDKKLKERL', 'ERTILSDFPQWWFASLDFLNC', 'ACDEFGHIKLMNPQRSTVWY']) >>> desc.formula(amide=True) >>> for v in desc.descriptor: ... print(v[0]) C122 H197 N35 O39 C121 H168 N28 O33 S C106 H157 N29 O30 S2
See also
New in version v2.7.6.
-
calculate_MW(amide=False, append=False)[source]¶ Method to calculate the molecular weight [g/mol] of every sequence in the attribute
sequences.- Parameters
amide – {boolean} whether the sequences are C-terminally amidated (subtracts 0.95 from the MW).
append – {boolean} whether the produced descriptor values should be appended to the existing ones in the attribute
descriptor.
- Returns
array of descriptor values in the attribute
descriptor- Example
>>> desc = GlobalDescriptor('IAESFKGHIPL') >>> desc.calculate_MW(amide=True) >>> desc.descriptor array([[ 1210.43]])
See also
Changed in version v2.1.5: amide option added
-
calculate_charge(ph=7.0, amide=False, append=False)[source]¶ Method to overall charge of every sequence in the attribute
sequences.The method used is first described by Bjellqvist. In the case of amidation, the value for the ‘Cterm’ pKa is 15 (and Cterm is added to the pos_pKs dictionary. The pKa scale is extracted from: http://www.hbcpnetbase.com/ (CRC Handbook of Chemistry and Physics, 96th ed).
pos_pKs = {‘Nterm’: 9.38, ‘K’: 10.67, ‘R’: 12.10, ‘H’: 6.04}
neg_pKs = {‘Cterm’: 2.15, ‘D’: 3.71, ‘E’: 4.15, ‘C’: 8.14, ‘Y’: 10.10}
- Parameters
ph – {float} ph at which to calculate peptide charge.
amide – {boolean} whether the sequences have an amidated C-terminus.
append – {boolean} whether the produced descriptor values should be appended to the existing ones in the attribute
descriptor.
- Returns
array of descriptor values in the attribute
descriptor- Example
>>> desc = GlobalDescriptor('KLAKFGKRSELVALSG') >>> desc.calculate_charge(ph=7.4, amide=True) >>> desc.descriptor array([[ 3.989]])
-
charge_density(ph=7.0, amide=False, append=False)[source]¶ Method to calculate the charge density (charge / MW) of every sequences in the attributes
sequences- Parameters
ph – {float} pH at which to calculate peptide charge.
amide – {boolean} whether the sequences have an amidated C-terminus.
append – {boolean} whether the produced descriptor values should be appended to the existing ones in the attribute
descriptor.
- Returns
array of descriptor values in the attribute
descriptor.- Example
>>> desc = GlobalDescriptor('GNSDLLIEQRTLLASDEF') >>> desc.charge_density(ph=6, amide=True) >>> desc.descriptor array([[-0.00097119]])
-
isoelectric_point(amide=False, append=False)[source]¶ Method to calculate the isoelectric point of every sequence in the attribute
sequences. The pK scale is extracted from: http://www.hbcpnetbase.com/ (CRC Handbook of Chemistry and Physics, 96th ed).pos_pKs = {‘Nterm’: 9.38, ‘K’: 10.67, ‘R’: 12.10, ‘H’: 6.04}
neg_pKs = {‘Cterm’: 2.15, ‘D’: 3.71, ‘E’: 4.15, ‘C’: 8.14, ‘Y’: 10.10}
- Parameters
amide – {boolean} whether the sequences have an amidated C-terminus.
append – {boolean} whether the produced descriptor values should be appended to the existing ones in the attribute
descriptor.
- Returns
array of descriptor values in the attribute
descriptor- Example
>>> desc = GlobalDescriptor('KLFDIKFGHIPQRST') >>> desc.isoelectric_point() >>> desc.descriptor array([[ 10.6796875]])
-
instability_index(append=False)[source]¶ Method to calculate the instability of every sequence in the attribute
sequences. The instability index is a prediction of protein stability based on the amino acid composition. ([1] K. Guruprasad, B. V Reddy, M. W. Pandit, Protein Eng. 1990, 4, 155–161.)- Parameters
append – {boolean} whether the produced descriptor values should be appended to the existing ones in the attribute
descriptor.- Returns
array of descriptor values in the attribute
descriptor- Example
>>> desc = GlobalDescriptor('LLASMNDLLAKRST') >>> desc.instability_index() >>> desc.descriptor array([[ 63.95714286]])
-
aromaticity(append=False)[source]¶ Method to calculate the aromaticity of every sequence in the attribute
sequences. According to Lobry, 1994, it is simply the relative frequency of Phe+Trp+Tyr.- Parameters
append – {boolean} whether the produced descriptor values should be appended to the existing ones in the attribute
descriptor.- Returns
array of descriptor values in the attribute
descriptor- Example
>>> desc = GlobalDescriptor('GLFYWRFFLQRRFLYWW') >>> desc.aromaticity() >>> desc.descriptor array([[ 0.52941176]])
-
aliphatic_index(append=False)[source]¶ Method to calculate the aliphatic index of every sequence in the attribute
sequences. According to Ikai, 1980, the aliphatic index is a measure of thermal stability of proteins and is dependant on the relative volume occupied by aliphatic amino acids (A,I,L & V). ([1] A. Ikai, J. Biochem. 1980, 88, 1895–1898.)- Parameters
append – {boolean} whether the produced descriptor values should be appended to the existing ones in the attribute
descriptor.- Returns
array of descriptor values in the attribute
descriptor- Example
>>> desc = GlobalDescriptor('KWLKYLKKLAKLVK') >>> desc.aliphatic_index() >>> desc.descriptor array([[ 139.28571429]])
-
boman_index(append=False)[source]¶ Method to calculate the boman index of every sequence in the attribute
sequences. According to Boman, 2003, the boman index is a measure for protein-protein interactions and is calculated by summing over all amino acid free energy of transfer [kcal/mol] between water and cyclohexane,[2] followed by dividing by sequence length. ([1] H. G. Boman, D. Wade, I. a Boman, B. Wåhlin, R. B. Merrifield, FEBS Lett. 1989, 259, 103–106. [2] A. Radzick, R. Wolfenden, Biochemistry 1988, 27, 1664–1670.)See also
- Parameters
append – {boolean} whether the produced descriptor values should be appended to the existing ones in the attribute
descriptor.- Returns
array of descriptor values in the attribute
descriptor- Example
>>> desc = GlobalDescriptor('GLFDIVKKVVGALGSL') >>> desc.boman_index() >>> desc.descriptor array([[-1.011875]])
-
hydrophobic_ratio(append=False)[source]¶ Method to calculate the hydrophobic ratio of every sequence in the attribute
sequences, which is the relative frequency of the amino acids A,C,F,I,L,M & V.- Parameters
append – {boolean} whether the produced descriptor values should be appended to the existing ones in the attribute
descriptor.- Returns
array of descriptor values in the attribute
descriptor- Example
>>> desc = GlobalDescriptor('VALLYWRTVLLAIII') >>> desc.hydrophobic_ratio() >>> desc.descriptor array([[ 0.73333333]])
-
calculate_all(ph=7.4, amide=True)[source]¶ Method combining all global descriptors and appending them into the feature matrix in the attribute
descriptorand corresponding feature names infeaturenames.- Parameters
ph – {float} pH at which to calculate peptide charge
amide – {boolean} whether the sequences have an amidated C-terminus.
- Returns
array of descriptor values in the attribute
descriptorand corresponding feature names in
featurenames:Example:>>> desc = GlobalDescriptor('AFGHFKLKKLFIFGHERT') >>> desc.calculate_all(amide=True) >>> desc.featurenames ['Length', 'MW', 'Charge', 'ChargeDensity', 'pI', 'InstabilityInd', 'Aromaticity', 'AliphaticInd', 'BomanInd', 'HydrophRatio'] >>> desc.descriptor array([[ 18., 2.17559000e+03, 1.87167619e-03, 1.16757812e+01, ... 1.10555556e+00, 4.44444444e-01]]) >>> desc.save_descriptor('/path/to/outputfile.csv') # save the descriptor data (with feature names header)
-
class
modlamp.descriptors.PeptideDescriptor(seqs, scalename='Eisenberg')[source]¶ Base class for peptide descriptors. The following amino acid descriptor scales are available for descriptor calculation:
AASI (An amino acid selectivity index scale for helical antimicrobial peptides, [1] D. Juretić, D. Vukicević, N. Ilić, N. Antcheva, A. Tossi, J. Chem. Inf. Model. 2009, 49, 2873–2882.)
ABHPRK (modlabs inhouse physicochemical feature scale (Acidic, Basic, Hydrophobic, Polar, aRomatic, Kink-inducer)
argos (Argos hydrophobicity amino acid scale, [2] Argos, P., Rao, J. K. M. & Hargrave, P. A., Eur. J. Biochem. 2005, 128, 565–575.)
bulkiness (Amino acid side chain bulkiness scale, [3] J. M. Zimmerman, N. Eliezer, R. Simha, J. Theor. Biol. 1968, 21, 170–201.)
charge_phys (Amino acid charge at pH 7.0 - Hystidine charge +0.1.)
charge_acid (Amino acid charge at acidic pH - Hystidine charge +1.0.)
cougar (modlabs inhouse selection of global peptide descriptors)
eisenberg (the Eisenberg hydrophobicity consensus amino acid scale, [4] D. Eisenberg, R. M. Weiss, T. C. Terwilliger, W. Wilcox, Faraday Symp. Chem. Soc. 1982, 17, 109.)
Ez (potential that assesses energies of insertion of amino acid side chains into lipid bilayers, [5] A. Senes, D. C. Chadi, P. B. Law, R. F. S. Walters, V. Nanda, W. F. DeGrado, J. Mol. Biol. 2007, 366, 436–448.)
flexibility (amino acid side chain flexibilitiy scale, [6] R. Bhaskaran, P. K. Ponnuswamy, Int. J. Pept. Protein Res. 1988, 32, 241–255.)
grantham (amino acid side chain composition, polarity and molecular volume, [8] Grantham, R. Science. 185, 862–864 (1974).)
gravy (GRAVY hydrophobicity amino acid scale, [9] J. Kyte, R. F. Doolittle, J. Mol. Biol. 1982, 157, 105–132.)
hopp-woods (Hopp-Woods amino acid hydrophobicity scale,*[10] T. P. Hopp, K. R. Woods, Proc. Natl. Acad. Sci. 1981, 78, 3824–3828.*)
ISAECI (Isotropic Surface Area (ISA) and Electronic Charge Index (ECI) of amino acid side chains, [11] E. R. Collantes, W. J. Dunn, J. Med. Chem. 1995, 38, 2705–2713.)
janin (Janin hydrophobicity amino acid scale, [12] J. L. Cornette, K. B. Cease, H. Margalit, J. L. Spouge, J. A. Berzofsky, C. DeLisi, J. Mol. Biol. 1987, 195, 659–685.)
kytedoolittle (Kyte & Doolittle hydrophobicity amino acid scale, [13] J. Kyte, R. F. Doolittle, J. Mol. Biol. 1982, 157, 105–132.)
levitt_alpha (Levitt amino acid alpha-helix propensity scale, extracted from http://web.expasy.org/protscale. [14] M. Levitt, Biochemistry 1978, 17, 4277-4285.)
MSS (A graph-theoretical index that reflects topological shape and size of amino acid side chains, [15] C. Raychaudhury, A. Banerjee, P. Bag, S. Roy, J. Chem. Inf. Comput. Sci. 1999, 39, 248–254.)
MSW (Amino acid scale based on a PCA of the molecular surface based WHIM descriptor (MS-WHIM), extended to natural amino acids, [16] A. Zaliani, E. Gancia, J. Chem. Inf. Comput. Sci 1999, 39, 525–533.)
pepArc (modlabs pharmacophoric feature scale, dimensions are: hydrophobicity, polarity, positive charge, negative charge, proline.)
pepcats (modlabs pharmacophoric feature based PEPCATS scale, [17] C. P. Koch, A. M. Perna, M. Pillong, N. K. Todoroff, P. Wrede, G. Folkers, J. A. Hiss, G. Schneider, PLoS Comput. Biol. 2013, 9, e1003088.)
polarity (Amino acid polarity scale, [18] J. M. Zimmerman, N. Eliezer, R. Simha, J. Theor. Biol. 1968, 21, 170–201.)
PPCALI (modlabs inhouse scale derived from a PCA of 143 amino acid property scales, [19] C. P. Koch, A. M. Perna, M. Pillong, N. K. Todoroff, P. Wrede, G. Folkers, J. A. Hiss, G. Schneider, PLoS Comput. Biol. 2013, 9, e1003088.)
refractivity (Relative amino acid refractivity values, [20] T. L. McMeekin, M. Wilensky, M. L. Groves, Biochem. Biophys. Res. Commun. 1962, 7, 151–156.)
t_scale (A PCA derived scale based on amino acid side chain properties calculated with 6 different probes of the GRID program, [21] M. Cocchi, E. Johansson, Quant. Struct. Act. Relationships 1993, 12, 1–8.)
TM_tend (Amino acid transmembrane propensity scale, extracted from http://web.expasy.org/protscale, [22] Zhao, G., London E. Protein Sci. 2006, 15, 1987-2001.)
z3 (The original three dimensional Z-scale, [23] S. Hellberg, M. Sjöström, B. Skagerberg, S. Wold, J. Med. Chem. 1987, 30, 1126–1135.)
z5 (The extended five dimensional Z-scale, [24] M. Sandberg, L. Eriksson, J. Jonsson, M. Sjöström, S. Wold, J. Med. Chem. 1998, 41, 2481–2491.)
Further, amino acid scale independent methods can be calculated with help of the
GlobalDescriptorclass.-
__init__(seqs, scalename='Eisenberg')[source]¶ - Parameters
seqs – a .fasta file with sequences, a list of sequences or a single sequence as string to calculate the descriptor values for.
scalename – {str} name of the amino acid scale (one of the given list above) used to calculate the descriptor values
- Returns
initialized attributes
sequences,namesand dictionaryscalewith amino acid scale values of the scale name inscalename.- Example
>>> AMP = PeptideDescriptor('KLLKLLKKLLKLLK','pepcats') >>> AMP.sequences ['KLLKLLKKLLKLLK'] >>> seqs = PeptideDescriptor('/Path/to/file.fasta', 'eisenberg') # load sequences from .fasta file >>> seqs.sequences ['AFDGHLKI','KKLQRSDLLRTK','KKLASCNNIPPR'...]
-
load_scale(scalename)[source]¶ Method to load amino acid values from a given scale
- Parameters
scalename – {str} name of the amino acid scale to be loaded.
- Returns
loaded amino acid scale values in a dictionary in the attribute
scale.
See also
-
calculate_autocorr(window, append=False)[source]¶ Method for auto-correlating the amino acid values for a given descriptor scale
- Parameters
window – {int} correlation window for descriptor calculation in a sliding window approach
append – {boolean} whether the produced descriptor values should be appended to the existing ones in the attribute
descriptor.
- Returns
calculated descriptor numpy.array in the attribute
descriptor.- Example
>>> AMP = PeptideDescriptor('GLFDIVKKVVGALGSL','PPCALI') >>> AMP.calculate_autocorr(7) >>> AMP.descriptor array([[ 1.28442339e+00, 1.29025116e+00, 1.03240901e+00, .... ]]) >>> AMP.descriptor.shape (1, 133)
Changed in version v.2.3.0.
-
calculate_crosscorr(window, append=False)[source]¶ Method for cross-correlating the amino acid values for a given descriptor scale
- Parameters
window – {int} correlation window for descriptor calculation in a sliding window approach
append – {boolean} whether the produced descriptor values should be appended to the existing ones in the attribute
descriptor.
- Returns
calculated descriptor numpy.array in the attribute
descriptor.- Example
>>> AMP = PeptideDescriptor('GLFDIVKKVVGALGSL','pepcats') >>> AMP.calculate_crosscorr(7) >>> AMP.descriptor array([[ 0.6875 , 0.46666667, 0.42857143, 0.61538462, 0.58333333, ... ]]) >>> AMP.descriptor.shape (1, 147)
-
calculate_moment(window=1000, angle=100, modality='max', append=False)[source]¶ Method for calculating the maximum or mean moment of the amino acid values for a given descriptor scale and window.
- Parameters
window – {int} amino acid window in which to calculate the moment. If the sequence is shorter than the window, the length of the sequence is taken. So if the default window of 1000 is chosen, for all sequences shorter than 1000, the global hydrophobic moment will be calculated. Otherwise, the maximal hydrophiobic moment for the chosen window size found in the sequence will be returned.
angle – {int} angle in which to calculate the moment. 100 for alpha helices, 180 for beta sheets.
modality – {‘all’, ‘max’ or ‘mean’} Calculate respectively maximum or mean hydrophobic moment. If all, moments for all windows are returned.
append – {boolean} whether the produced descriptor values should be appended to the existing ones in the attribute
descriptor.
- Returns
Calculated descriptor as a numpy.array in the attribute
descriptorand all possible global values inall_moms(needed for thecalculate_profile()method)- Example
>>> AMP = PeptideDescriptor('GLFDIVKKVVGALGSL', 'eisenberg') >>> AMP.calculate_moment() >>> AMP.descriptor array([[ 0.48790226]])
-
calculate_global(window=1000, modality='max', append=False)[source]¶ Method for calculating a global / window averaging descriptor value of a given AA scale
- Parameters
window – {int} amino acid window in which to calculate the moment. If the sequence is shorter than the window, the length of the sequence is taken.
modality – {‘max’ or ‘mean’} Calculate respectively maximum or mean hydrophobic moment.
append – {boolean} whether the produced descriptor values should be appended to the existing ones in the attribute
descriptor.
- Returns
Calculated descriptor as a numpy.array in the attribute
descriptorand all possible global values inall_globs(needed for thecalculate_profile()method)- Example
>>> AMP = PeptideDescriptor('GLFDIVKKVVGALGSL','eisenberg') >>> AMP.calculate_global(window=1000, modality='max') >>> AMP.descriptor array([[ 0.44875]])
-
calculate_profile(prof_type='uH', window=7, append=False)[source]¶ Method for calculating hydrophobicity or hydrophobic moment profiles for given sequences and fitting for slope and intercept. The hydrophobicity scale used is “eisenberg”
- Parameters
prof_type – prof_type of profile, available: ‘H’ for hydrophobicity or ‘uH’ for hydrophobic moment
window – {int} size of sliding window used (odd-numbered).
append – {boolean} whether the produced descriptor values should be appended to the existing ones in the attribute
descriptor.
- Returns
Fitted slope and intercept of calculated profile for every given sequence in the attribute
descriptor.- Example
>>> AMP = PeptideDescriptor('KLLKLLKKVVGALG','kytedoolittle') >>> AMP.calculate_profile(prof_type='H') >>> AMP.descriptor array([[ 0.03731293, 0.19246599]])
-
calculate_arc(modality='max', append=False)[source]¶ Method for calculating property arcs as seen in the helical wheel plot. Use for binary amino acid scales only.
- Parameters
modality – modality of the arc to calculate, to choose between “max” and “mean”.
append – if true, append to current descriptor stored in the descriptor attribute.
- Returns
calculated descriptor as numpy.array in the descriptor attribute.
- Example
>>> arc = PeptideDescriptor("KLLKLLKKLLKLLK", scalename="peparc") >>> arc.calculate_arc(modality="max", append=False) >>> arc.descriptor array([[200, 160, 160, 0, 0]])
2.2. modlamp.sequences¶
This module incorporates different classes to generate peptide sequences with different characteristics from scratch. The following classes are available:
Class |
Characteristics |
|---|---|
Generates random sequences with a specified amino acid distribution. |
|
Generates presumed amphipathic helical sequences with a hydrophobic moment. |
|
Generates presumed amphipathic helices with a kink (Pro residue). |
|
Generates presumed oblique oriented sequences in presence of libid membranes. |
|
Generates centrosymmetric sequences with a symmetry axis. |
|
Generates presumed amphipathic helices with controlled hydrophobic arc size. |
|
Generates sequences with the amino acid probabiliy of helical ACPs. |
|
Generates a mixed library of sequences of most other classes. |
|
Generates presumed amphipathic helices with a heparin-binding-domain. |
|
Generates sequences from most frequent ngrams in the APD3. |
All classes share the same method generate_sequences() for sequence generation, potentially enabling
automation or looping over different classes.
Note
During the process of sequence generation, duplicates are only removed for the MixedLibrary
class. To remove duplicates, call the class methods self.filter_duplicates().
See also
modlamp.core.BaseSequence from which all classes in this module inherit.
-
class
modlamp.sequences.Random(seqnum, lenmin=7, lenmax=28)[source]¶ Class for random peptide sequences.
This class incorporates methods for generating peptide random peptide sequences of defined length. The amino acid probabilities can be chosen from different probabilities:
rand: equal probabilities for all amino acids
AMP: amino acid probabilities taken from the antimicrobial peptide database APD3, March 17, 2016, containing 2674 sequences.
AMPnoCM: same amino acid probabilities as AMP but lacking Cys and Met (for synthesizability)
ACP: amino acid probabilities taken from 339 linear peptides in CancerPPD database CancerPPD <http://http://crdd.osdd.net/raghava/cancerppd/>
randnoCM: equal probabilities for all amino acids, except 0.0 for both Cys and Met (for synthesizability)
The probability values for all natural AA can be found in the following table:
AA
rand
AMP
AMPnoCM
randnoCM
ACP
A
0.05
0.0766
0.0812275
0.05555555
0.14526966
C
0.05
0.071
0.0
0.0
D
0.05
0.026
0.0306275
0.05555555
0.00690031
E
0.05
0.0264
0.0310275
0.05555555
0.00780824
F
0.05
0.0405
0.0451275
0.05555555
0.06991102
G
0.05
0.1172
0.1218275
0.05555555
0.04957327
H
0.05
0.021
0.0256275
0.05555555
0.01725077
I
0.05
0.061
0.0656275
0.05555555
0.05647358
K
0.05
0.0958
0.1004275
0.05555555
0.27637552
L
0.05
0.0838
0.0884275
0.05555555
0.17759216
M
0.05
0.0123
0.0
0.0
0.00998729
N
0.05
0.0386
0.0432275
0.05555555
0.00798983
P
0.05
0.0463
0.0509275
0.05555555
0.01307427
Q
0.05
0.0251
0.0297275
0.05555555
0.00381333
R
0.05
0.0545
0.0591275
0.05555555
0.02941711
S
0.05
0.0613
0.0659275
0.05555555
0.02651171
T
0.05
0.0455
0.0501275
0.05555555
0.01543490
V
0.05
0.0572
0.0618275
0.05555555
0.04013074
W
0.05
0.0155
0.0201275
0.05555555
0.04067550
Y
0.05
0.0244
0.0290275
0.05555555
0.00581079
-
generate_sequences(proba='rand')[source]¶ Method to actually generate the sequences.
- Parameters
proba – {str or list} AA probability to be used to generate sequences. Available from str: AMP, AMPnoCM, rand, randnoCM. You can also provide your own list of porbabilities as a list (in AA order, length 20, sum to 1)
- Returns
A list of random AMP sequences with defined AA probabilities
- Example
>>> b = Random(6, 5, 20) >>> b.generate_sequences(proba='AMP') >>> b.sequences ['CYGALWHIFV','NIVRHHAPSTVIK','LCPNPILGIV','TAVVRGKESLTP','GTGSVCKNSCRGRFGIIAF','VIIGPSYGDAEYA']
-
class
modlamp.sequences.Helices(seqnum, lenmin=7, lenmax=28)[source]¶ Class for peptide sequences probable to form helices. This class incorporates methods for generating presumed amphipathic alpha-helical peptide sequences. These sequences are generated by placing basic residues along the sequence with distance 3-4 AA to each other. The remaining empty spots are filled up by hydrophobic AAs.
-
generate_sequences()[source]¶ Method to generate amphipathic helical sequences with class features defined in
Helices()- Returns
In the attribute
sequences: a list of sequences with presumed amphipathic helical structure.- Example
>>> h = Helices(5, 7, 21) >>> h.generate_sequences() >>> h.sequences ['KGIKVILKLAKAGVKAVRL','IILKVGKV','IAKAGRAIIK','LKILKVVGKGIRLIVRIIKAL','KAGKLVAKGAKVAAKAIKI']
-
-
class
modlamp.sequences.Kinked(seqnum, lenmin=7, lenmax=28)[source]¶ Class for peptide sequences probable to form helices with a kink.
This class incorporates methods for presumed kinked amphipathic alpha-helical peptide sequences: Sequences are generated by placing basic residues along the sequence with distance 3-4 AA to each other. The remaining spots are filled up by hydrophobic AAs. Then, a basic residue is replaced by proline, presumably leading to a kink in the hydrophobic face of the amphipathic helices.
-
generate_sequences()[source]¶ Method to actually generate the presumed kinked sequences with features defined in the class instances.
- Returns
sequence list with strings stored in the attribute
sequences- Example
>>> k = Kinked(8, 7, 28) >>> k.generate_sequences() >>> k.sequences ['IILRLHPIG','ARGAKVAIKAIRGIAPGGRVVAKVVKVG','GGKVGRGVAFLVRIILK','KAVKALAKGAPVILCVAKVI', ...]
-
-
class
modlamp.sequences.Oblique(seqnum, lenmin=7, lenmax=28)[source]¶ Class for oblique sequences with a so called linear hydrophobicity gradient.
This class incorporates methods for generating peptide sequences with a linear hydrophobicity gradient, meaning that these sequences have a hydrophobic tail. This feature gives rise to the hypothesis that they orient themselves tilted/oblique in membrane environment.
-
generate_sequences()[source]¶ Method to generate the possible oblique sequences.
- Returns
A list of sequences in the attribute
sequences.- Example
>>> o = Oblique(4, 10, 30) >>> o.generate_sequences() >>> o.sequences ['GLLKVIRIAAKVLKVAVLVGIIAI','AIGKAGRLALKVIKVVIKVALILLAAVA','KILRAAARVIKGGIKAIVIL','VRLVKAIGKLLRIILRLARLAVGGILA']
-
-
class
modlamp.sequences.Centrosymmetric(seqnum, lenmin=7, lenmax=28)[source]¶ Class for peptide sequences produced out of 7 AA centro-symmetric blocks yielding peptides of length 14 or 21 AA (2*7 or 3*7).
This class incorporates methods to generate special peptide sequences with an overall presumed hydrophobic moment. Sequences are generated by centro-symmetric blocks of seven amino acids. Two or three blocks are added to build a final sequence of length 14 or 21 amino acids length. If the option
symmetricis used, two or three identical blocks are concatenated. If the the optionasymmetricis used, two or three different blocks are concatenated.-
generate_sequences(symmetry='asymmetric')[source]¶ Generate overall symmetric or asymmetric sequences out of two or three blocks of centro-symmetric blocks of 7 amino acids. The resulting sequence presumably has a large hydrophobic moment.
- Parameters
symmetry – {str} type of centrosymmetric sequences.
symmetric: builds sequences out of only one block,asymmetric: builds sequences out of different blocks- Returns
In the attribute
sequences: centro-symmetric peptide sequences of the form [h,+,h,a,h,+,h] with h = hydrophobic AA, + = basic AA, a = anchor AA (F,Y,W,(P)), sequence length is 14 or 21 AA- Example
>>> s = Centrosymmetric(5) >>> s.generate_sequences(symmetry='symmetric') >>> s.sequences ['ARIFIRAARIFIRA','GRIYIRGGRIYIRGGRIYIRG','IRGFGRIIRGFGRIIRGFGRI','GKAYAKGGKAYAKG','AKGYGKAAKGYGKAAKGYGKA']
-
-
class
modlamp.sequences.AmphipathicArc(seqnum, lenmin=7, lenmax=28)[source]¶ Class for generating positively-charged amphipathic peptide sequences based on an alpha-helix pattern with different arc sizes.
The probability values for the Hydrophobic and Polar positions of the helix can be found in the following table:
AA
Hydr
Polar
A
0.00
0.045
C
0.00
0.00
D
0.00
0.045
E
0.00
0.045
F
0.16
0.00
G
0.00
0.045
H
0.00
0.045
I
0.16
0.00
K
0.00
0.25
L
0.16
0.00
M
0.00
0.00
N
0.00
0.05
P
0.00
0.045
Q
0.00
0.045
R
0.00
0.25
S
0.00
0.045
T
0.00
0.045
V
0.16
0.00
W
0.16
0.00
Y
0.16
0.045
-
generate_sequences(arcsize=180)[source]¶ Method to generate the possible amphipathic helices with defined hydrophobic arc sizes (option
arcsize), with mixed arc sizes (optionarcsize=None)- Parameters
arcsize – {int} to choose among 100, 140, 180, 220, 260, or choose mixed to generate a mixture.
- Returns
A list of sequences in the attribute
sequences.- Example
>>> from modlamp.sequences import AmphipathicArc >>> amphi_hel = AmphipathicArc(4, 10, 25) >>> amphi_hel.generate_sequences(100) >>> amphi_hel.sequences ['YLYANLRQE', 'GVKPRIK', 'RWKKKVKDSVKDFEKRFKDIEKRIQRKLA', 'KIKEQLRNSVSGWHRN']
-
make_H_gradient()[source]¶ Method to mutate the generated sequences to have a hydrophobic gradient by substituting the last third of the sequence amino acids to hydrophobic.
- Returns
A list of sequences in
sequences- Example
>>> amphi_grad = AmphipathicArc(10, 7, 30) >>> amphi_grad.generate_sequences(180) >>> amphi_grad.make_H_gradient() >>> amphi_grad.sequences
-
-
class
modlamp.sequences.HelicesACP(seqnum, lenmin=7, lenmax=28)[source]¶ Class for peptides sequences with the amino acid probability of alpha-helical ACPs.
This class incorporates methods for generating presumed alpha-helical peptides with the amino acid probability distribution of alpha-helical ACPs. For each of the positions in the helix (1-18) the amino acid distribution among 62 anuran and hymenopteran alpha-helical ACPs was computed and is used to design the new sequences (G. Gabernet, A. T. Müller, J. A. Hiss, G. Schneider, Med. Chem. Commun. 2016, 7, 2232–2245.).
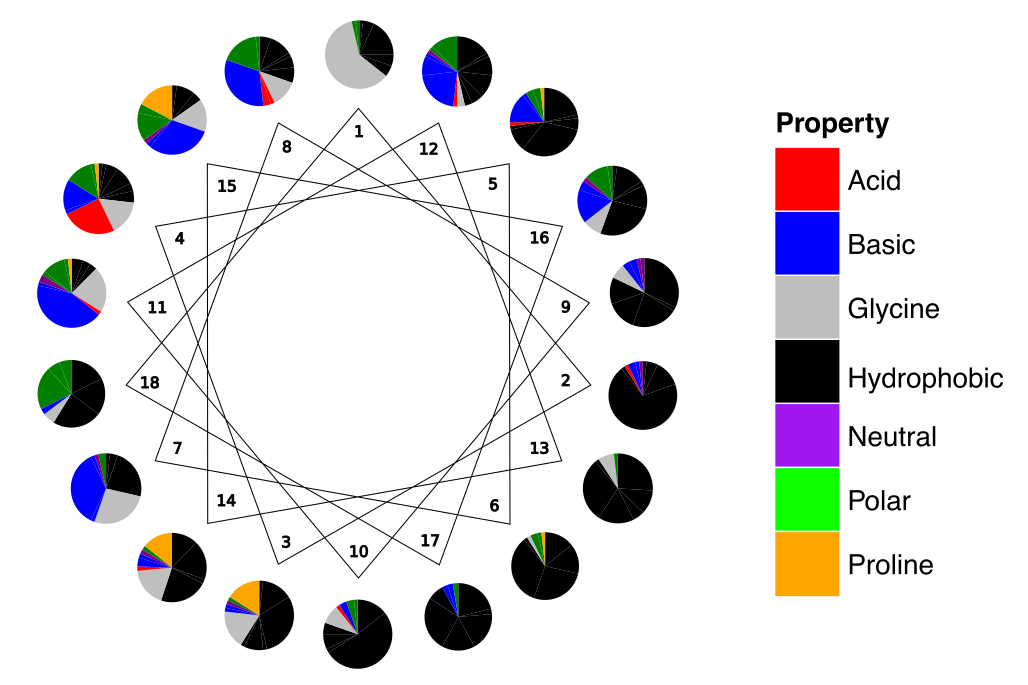
-
generate_sequences()[source]¶ Method to generate the sequences with the mentioned amino acid probabilities.
- Returns
A list of potentially helical peptides with the amino acid distribution of ACP helical peptides according to the position in the helix wheel.
- Example
>>> from modlamp.sequences import HelicesACP >>> helACP = HelicesACP(4, 7, 18) >>> helACP.generate_sequences() >>> helACP.sequences ['FLFDVAKKVAGTALT', 'GLGIILGAGG', 'GLRIKLGVWAKKA', 'GFWGFIKTI']
-
-
class
modlamp.sequences.MixedLibrary(seqnum, centrosymmetric=1, centroasymmetric=1, helix=1, kinked=1, oblique=1, rand=1, randAMP=1, randAMPnoCM=1)[source]¶ Class for holding a virtual peptide library.
This class
MixedLibraryincorporates methods to generate a virtual peptide library composed out of different sub-libraries. The available library subtypes are all from the classesCentrosymmetric,Helices,Kinked,ObliqueorRandom.-
__init__(seqnum, centrosymmetric=1, centroasymmetric=1, helix=1, kinked=1, oblique=1, rand=1, randAMP=1, randAMPnoCM=1)[source]¶ initializing method of the class
MixedLibrary. Except from number, all other parameters are ratios of sequences of the given sequence class.- Parameters
seqnum – {int} number of sequences to be generated
centrosymmetric – ratio of symmetric centrosymmetric sequences in the library
centroasymmetric – ratio of asymmetric centrosymmetric sequences in the library
helix – ratio of amphipathic helical sequences in the library
kinked – ratio of kinked amphipathic helical sequences in the library
oblique – ratio of oblique oriented amphipathic helical sequences in the library
rand – ratio of random sequneces in the library
randAMP – ratio of random sequences with APD2 amino acid distribution in the library
randAMPnoCM – ratio of random sequences with APD2 amino acid distribution without Cys and Met in the library
Warning
If duplicate sequences are created, these are removed during the creation process. It is therefore quite probable that you will not get the exact size of library that you entered as the parameter number. If you generate a small library, it can also happen that the size is bigger than expected, because ratios are rounded.
-
generate_sequences()[source]¶ This method generates a virtual sequence library with the subtype ratios initialized in class
MixedLibrary(). All sequences are between 7 and 28 amino acids in length.- Returns
a virtual library of sequences in the attribute
sequences, the sub-library class names innames, the number of sequences generated for each class innumsand the library size inlibsize.- Example
>>> lib = MixedLibrary(10000, centrosymmetric=5, centroasymmetric=5, helix=3, kinked=3, oblique=2, rand=10, randAMP=10,randAMPnoCM=5) >>> lib.generate_sequences() >>> lib.libsize # as duplicates were present, the library does not have the size that was sepecified 9126 >>> lib.sequences ['RHTHVAGSWYGKMPPSPQTL','MRIKLRKIPCILAC','DGINKEVKDSYGVFLK','LRLYLRLGRVWVRG','GKLFLKGGKLFLKGGKLFLKG',...] >>> lib.nums {'AMP': 2326, 'asy': 1163, 'hel': 698, 'knk': 698, 'nCM': 1163, 'obl': 465, 'ran': 2326, 'sym': 1163}
-
-
class
modlamp.sequences.Hepahelices(seqnum, lenmin=7, lenmax=28)[source]¶ Class for peptide sequences probable to form helices and include a heparin-binding-domain.
This class is used to construct presumed amphipathic helices that include a heparin-binding-domain (HBD) probable to bind heparin. The HBD sequence for alpha-helices usually has the following form: XBBBXXBX (B: basic AA; X: hydrophobic, uncharged AA, with mainly Ser & Gly).
- Reference
Munoz, E. M. & Linhardt, R. J. Heparin-Binding Domains in Vascular Biology. Arterioscler. Thromb. Vasc. Biol. 24, 1549–1557 (2004).
New in version v2.3.1.
-
generate_sequences()[source]¶ Method to generate helical sequences with class features defined in
Hepahelices()- Returns
In the attribute
sequences: a list of sequences including a heparin-binding-domain.- Example
>>> h = Hepahelices(10, 8,21) # minimal length: 8, maximal length: 50 >>> h.generate_sequences() >>> h.sequences ['GRLARSLKRKLNRLVRGGGRLVRGGG', 'IRSIRRRLSKLARSLGRGARSLGRG', 'RAVKRKVNKLLKGAAKVLKGAAKVLKGAAK', ... ]
-
class
modlamp.sequences.AMPngrams(seqnum, n_min=3, n_max=11)[source]¶ Class for sequence generation from the most prominent ngrams (2, 3, 4grams) found in all natural AMP sequences extracted from the APD3 (version August 2016 with 2727 sequences). For all 2, 3 and 4grams, all possible ngrams were generated from all sequences and the top 50 most frequent assembled into a list. Finally, leading and tailing spaces were striped and duplicates as well as ngrams containing spaces were removed.
New in version v2.4.1.
See also
-
__init__(seqnum, n_min=3, n_max=11)[source]¶ - Parameters
seqnum – {int} number of sequences to be generated
n_min – {int} minimum number of ngrams to take for sequence assembly
n_max – {int} maximum number of ngrams to take for sequence assembly
- Example
>>> s = AMPngrams(10) >>> s.generate_sequences() >>> s.sequences ['LAKSLGAGKYGGGKA', 'KAALESCVGGGGC', 'GCSGKAAAAAVG', 'GAASCKPCGEAKGLKVCY', 'IGGGCKITGESCVAGLWCGESTCGCSG', ...] >>> s.ngrams array(['AGK', 'CKI', 'RR', 'YGGG', 'LSGL', 'RG', 'YGGY', 'PRP', 'LGGG', ...]
-
2.3. modlamp.database¶
This module incorporates functions to connect to several peptide databases. It also allows to connect to a custom SQL database for which the configuration is given in a specified config file.
-
modlamp.database.query_database(table, columns=None, configfile='./modlamp/data/db_config.json')[source]¶ This function extracts experimental results from the modlab peptide database. All data from the given table and column names is extracted and returned.
- Parameters
table – the mysql database table to be queried
columns – a list of the column names {str} to be extracted from the table default:
*(all columns)configfile – location of the database configuration file containing the hostname etc. for the database to be queried.
- Returns
{numpy.array} queried data
- Example
>>> data = query_database(table='modlab_experiments', columns=['sequence', 'MCF7_activity', 'Saureus_activity']) Password: ********* Connecting to MySQL database... connection established! >>> data[:5] array([ ['ILGTILGILKGL', None, 1.0], ['ILGTILGFLKGL', None, 1.0], ['ILGNILGFLKGL', None, 1.0], ['ILGQILGILKGL', None, 1.0], ['ILGHILGYLKGL', None, 1.0]], dtype=object)
Note
If
NoneorNULLappears as a value, this means no data was measured for this peptide and not that activity is none (inactive).
-
modlamp.database.query_apd(ids)[source]¶ A function to query sequences from the antimicrobial peptide database APD. If the whole database should be scraped, simply look up the latest entry ID and take a
range(1, 'latestID')as function input.- Parameters
ids – {list of int} list of APD IDs to be queried from the database
- Returns
list of peptide sequences corresponding to entered ids.
- Example
>>> query_apd([15, 16, 18, 19, 20]) ['GLFDIVKKVVGALGSL', 'GLFDIVKKVVGAIGSL', 'GLFDIVKKVVGAFGSL', 'GLFDIAKKVIGVIGSL', 'GLFDIVKKIAGHIAGSI']
-
modlamp.database.query_camp(ids)[source]¶ A function to query sequences from the antimicrobial peptide database CAMP. If the whole database should be scraped, simply look up the latest entry ID and take a
range(1, 'latestID')as function input.- Parameters
ids – {list of int} list of CAMP IDs to be queried from the database
- Returns
list of peptide sequences corresponding to entered ids.
- Example
>>> query_camp([2705, 2706]) ['GLFDIVKKVVGALGSL', 'GLFDIVKKVVGTLAGL']
2.4. modlamp.datasets¶
This module incorporates functions to load different peptide datasets used for classification.
Function |
Data |
|---|---|
Antimicrobial peptides versus trans-membrane sequences |
|
AMPs from the APD3 versus other peptides from the UniProt database |
|
Anticancer peptides (CancerPPD) versus helical transmembrane sequences |
|
Anticancer peptides (CancerPPD) versus random scrambled AMP sequences |
|
A custom data set provided in |
-
class
modlamp.datasets.Bunch(**kwargs)[source]¶ Container object for datasets
Dictionary-like object that exposes its keys as attributes. Taken from the sklearn package.
- Example
>>> b = Bunch(a=1, b=2) >>> b['b'] 2 >>> b.b # key can also be called as attribute 2 >>> b.a = 3 >>> b['a'] 3 >>> b.c = 6 >>> b['c'] 6
-
modlamp.datasets.load_AMPvsTM()[source]¶ Function to load a dataset consisting of AMP sequences and transmembrane regions of proteins for classification.
The AMP class consists of an intersection of all activity annotations of the APD2 and CAMP databases, where for gram positive, gram negative and antifungal exact matches were observed. A detailed description of how the dataset was compiled can be found in the following publication: Schneider, P., Müller, A. T., Gabernet, G., Button, A. L., Posselt, G., Wessler, S., Hiss, J. A. and Schneider, G. (2016), Hybrid Network Model for “Deep Learning” of Chemical Data: Application to Antimicrobial Peptides. Mol. Inf.. doi:10.1002/minf.201600011
Classes
2
Samples per class
206
Samples total
412
Dimensionality
1
- Returns
Bunch, a dictionary-like object, the interesting attributes are:
sequences, the sequences,target, the classification labels,target_names, the meaning of the labels andfeature_names, the meaning of the features.- Example
>>> from modlamp.datasets import load_AMPvsTM >>> data = load_AMPvsTM() >>> data.sequences ['AAGAATVLLVIVLLAGSYLAVLA','LWIVIACLACVGSAAALTLRA','FYRFYMLREGTAVPAVWFSIELIFGLFA','GTLELGVDYGRAN',...] >>> list(data.target_names) ['TM', 'AMP'] >>> len(data.sequences) 412 >>> data.target[:10] array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0])
-
modlamp.datasets.load_AMPvsUniProt()[source]¶ Function to load a dataset consisting of the whole APD3 versus the same number of sequences randomly extracted from the UniProt database, to be used for classification.
The AMP class consists of 2600 AMP sequences from the APD3 (extracted Jan. 2016). The UniProt class consists of 2600 randomly extracted protein sequences from the UniProt Database with the search query length 10 TO 50 filtered for unnatural amino acids.
Classes
2
AMP Samples
2600
UniProt Samples
2600
Samples total
5200
Dimensionality
1
- Returns
Bunch, a dictionary-like object, the interesting attributes are:
sequences, the sequences,target, the classification labels,target_names, the meaning of the labels andfeature_names, the meaning of the features.- Example
>>> from modlamp.datasets import load_AMPvsUniProt >>> data = load_AMPvsUniProt() >>> data.sequences[:10] ['GLWSKIKEVGKEAAKAAAKAAGKAALGAVSEAV', 'YVPLPNVPQPGRRPFPTFPGQGPFNPKIKWPQGY', ... ] >>> list(data.target_names) ['AMP', 'UniProt'] >>> len(data.sequences) 5200 >>> data.target[:10] array([1, 1, 1, 1, 1, 1, 1, 1, 1, 1])
-
modlamp.datasets.load_ACPvsTM()[source]¶ Function to load a dataset consisting of ACP sequences from the CancerPPD database and negative peptides extracted from alpha-helical transmembrane regions of proteins for classification.
The ACP class consists of a collection of 413 ACPs from the CancerPPD database with length between 7 and 30 aa and without cysteines to facilitate peptide synthesis.
The Negative peptide set contains a random selection of 413 transmembrane alpha-helices (extracted from the PDBTM ) isolated directly from the proteins crystal structure.
Classes
2
ACP peptides
413
Negative peptides
413
Total peptides
826
Dimensionality
1
- Returns
Bunch, a dictionary-like object, the interesting attributes are:
sequences, the sequences,target, the classification labels,target_names, the meaning of the labels andfeature_names, the meaning of the features.- Example
>>> from modlamp.datasets import load_ACPvsTM >>> data = load_ACPvsTM() >>> data.sequences[:4] ['AAKKWAKAKWAKAKKWAKAA', 'AAVPIVNLKDELLFPSWEALFSGSE', 'AAWKWAWAKKWAKAKKWAKAA', 'AFGMALKLLKKVL'] >>> list(data.target_names) ['TM', 'ACP'] >>> len(data.sequences) 826
-
modlamp.datasets.load_ACPvsRandom()[source]¶ - Function to load a dataset consisting of ACP sequences from the CancerPPD database and negative peptides generated
randomly with the amino acid composition of AMPs.
The ACP class consists of a collection of 413 ACPs from the CancerPPD database with length between 7 and 30 aa and without cysteines to facilitate peptide synthesis.
The Negative peptide set contains a random selection of 413 randomly generated peptides with the amino acid composition of AMPs in the APD2 database.
Classes
2
ACP peptides
413
Negative peptides
413
Total peptides
826
Dimensionality
1
- Returns
Bunch, a dictionary-like object, the interesting attributes are:
sequences, the sequences,target, the classification labels,target_names, the meaning of the labels andfeature_names, the meaning of the features.- Example
>>> from modlamp.datasets import load_ACPvsRandom >>> data = load_ACPvsRandom() >>> data.sequences[:3] ['AAKKWAKAKWAKAKKWAKAA', 'AAVPIVNLKDELLFPSWEALFSGSE', 'AAWKWAWAKKWAKAKKWAKAA'] >>> list(data.target_names) ['Random', 'ACP'] >>> len(data.sequences) 826
-
modlamp.datasets.load_custom(filename)[source]¶ Function to load a custom dataset saved in
modlamp/data/as a.csvfile.The following header needs to be included: Nr. of sequences, Nr. of columns - 1, Class name for 0, Class name for 1
Example
.csvfile structure:4, 1, TM, AMP GTLEFDVTIGRAN, 0 GSNVHLASNLLA, 0 GLFDIVKKVVGALGSL, 0 GLFDIIKKIAESF, 0
- Parameters
filename – {str} filename of the data file to be loaded; the file must be located in
modlamp/data/- Returns
Bunch, a dictionary-like object, the interesting attributes are:
sequences, the sequences,target, the classification labels,target_names, the meaning of the labels andfeature_names, the meaning of the features.- Example
>>> from modlamp.datasets import load_custom >>> data = load_custom('custom_data.csv')
2.5. modlamp.plot¶
This module incorporates functions to plot different feature plots. The following functions are available:
Function |
Characteristics |
|---|---|
Generate a box plot for visualizing the distribution of a given feature. |
|
Generate a 2D scatter plot of 2 given features. |
|
Generate a 3D scatter plot of 3 given features. |
|
Generates a profile plot of a sequence to visualize potential linear gradients. |
|
Generates a helical wheel projection plot of a given sequence. |
|
Generates a probability density estimation plot of given data arrays. |
|
Generates a violin plot for given classes and corresponding distributions. |
|
Generates an amino acid frequency plot for all 20 natural amino acids. |
-
modlamp.plot.plot_feature(y_values, targets=None, y_label='feature values', x_tick_labels=None, filename=None, colors=None)[source]¶ Function to generate a box plot of 1 given feature. The different target classes given in targets are plottet as separate boxes.
- Parameters
y_values – Array of feature values to be plotted.
targets – List of target class values [string/binary] for the given feature data.
y_label – Axis label.
x_tick_labels – list of labels to be assigned to the ticks on the x-axis. Must match the number of targets.
filename – filename where to safe the plot. default = None
colors – {list} colors to take for plotting (strings in HEX formats).
- Returns
A feature box plot.
- Example
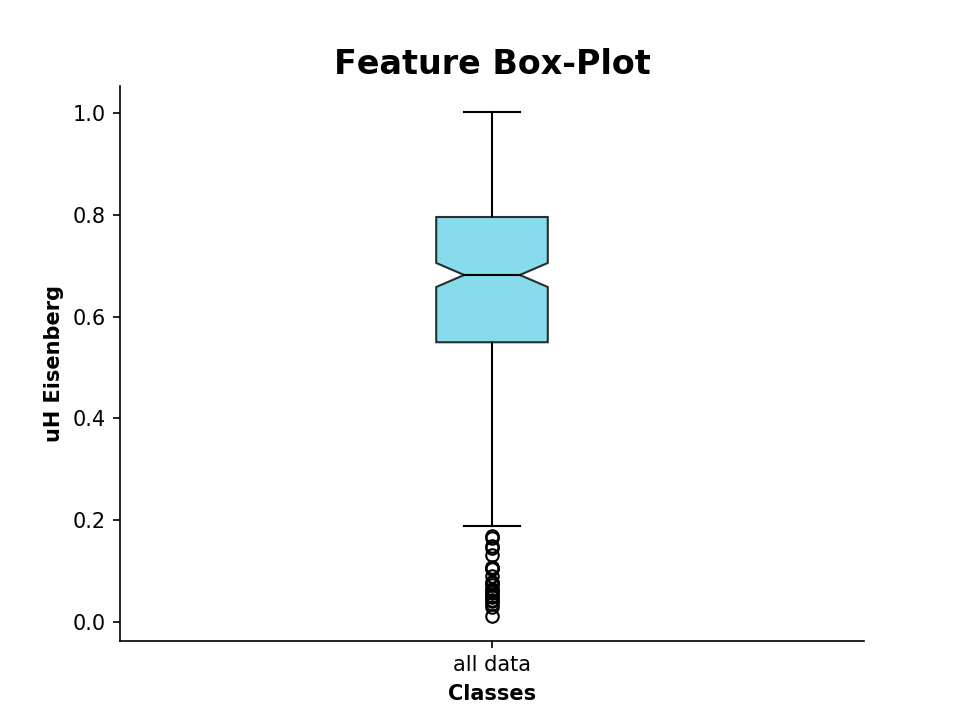
>>> plot_feature(desc.descriptor,y_label='uH Eisenberg') # desc: PeptideDescriptor instance
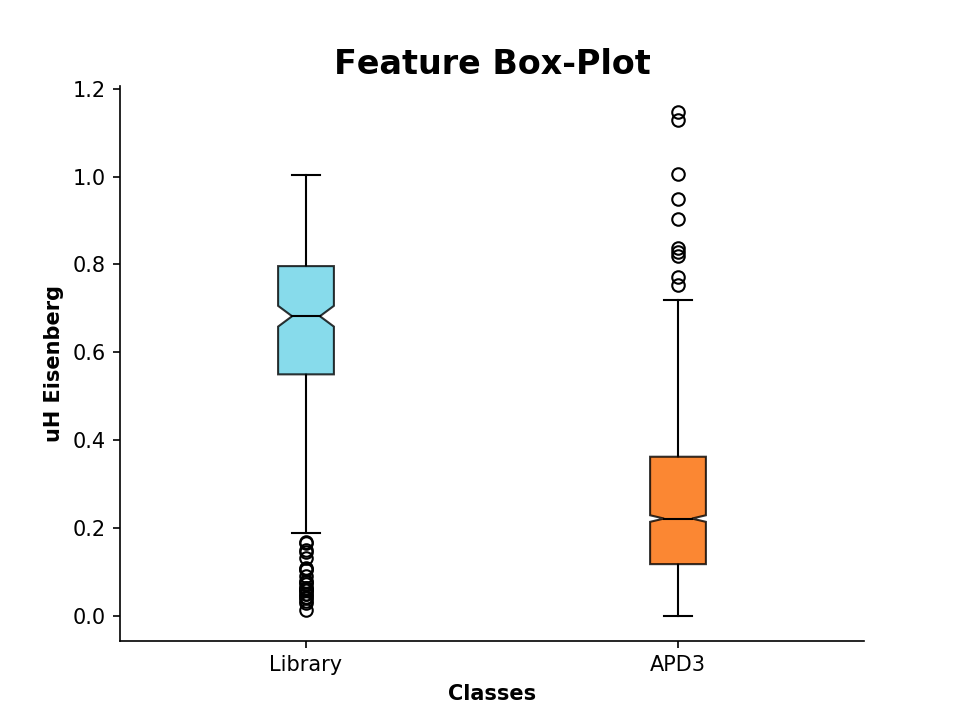
The same procedure also works for comparing two data sets:
>>> plot_feature((p.descriptor, apd.descriptor), y_label='uH Eisenberg', x_tick_labels=['Library', 'APD3'])
-
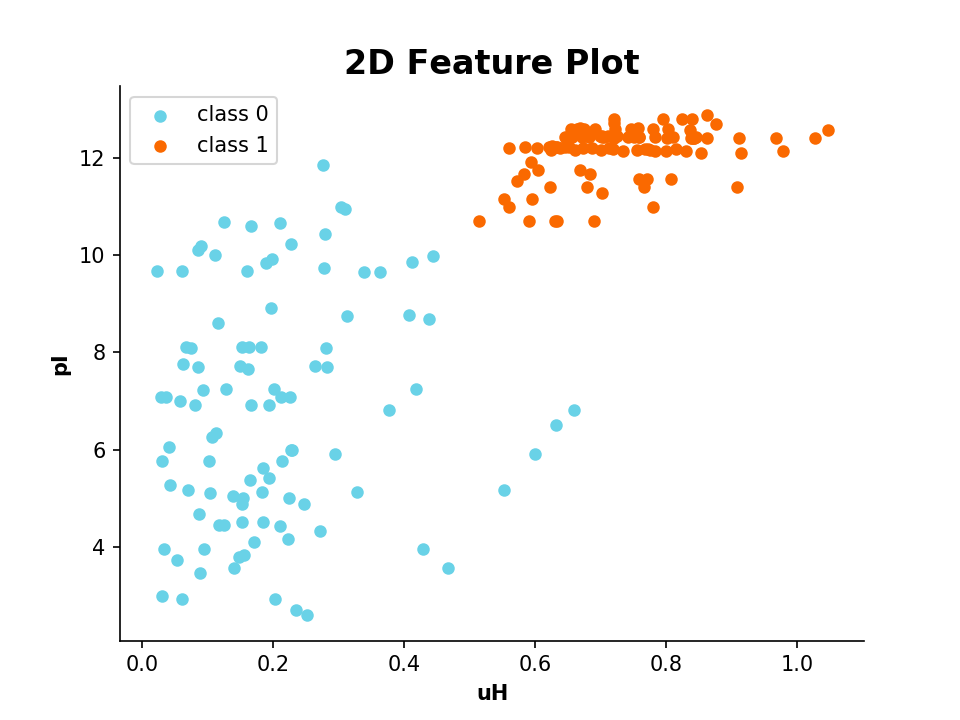
modlamp.plot.plot_2_features(x_values, y_values, targets=None, x_label='', y_label='', filename=None, colors=None)[source]¶ Function to generate a feature scatter plot of 2 given features. The different target classes given in targets are plottet in different colors.
- Parameters
x_values – Array of values of the feature to be plotted on the x-axis.
y_values – Array of values of the feature to be plotted on the y-axis.
targets – List of target class values [string/binary] for the given feature data.
x_label – X-axis label.
y_label – Y-axis label.
filename – filename where to safe the plot. default = None
colors – {list} colors to take for plotting (strings in HEX formats).
- Returns
A 2D feature scatter plot.
- Example
>>> plot_2_features(a.descriptor,b.descriptor,x_label='uH',y_label='pI',targets=targs)
-
modlamp.plot.plot_3_features(x_values, y_values, z_values, targets=None, x_label='', y_label='', z_label='', filename=None, colors=None)[source]¶ Function to generate a 3D feature scatter plot of 3 given features. The different target classes given in targets are plottet in different colors.
- Parameters
x_values – Array of values of the feature to be plotted on the x-axis.
y_values – Array of values of the feature to be plotted on the y-axis.
z_values – Array of values of the feature to be plotted on the z-axis.
targets – List of target class values {string/binary} for the given feature data.
x_label – {str} X-axis label.
y_label – {str} Y-axis label.
z_label – {str} Z-axis label.
filename – {str} filename where to safe the plot. default = None -> show the plot
colors – {list} colors to take for plotting (strings in HEX formats).
- Returns
A 3D feature scatter plot.
- Example
>>> plot_3_features(a.descriptor,b.descriptor,c.descriptor,x_label='uH',y_label='pI',z_label='length')
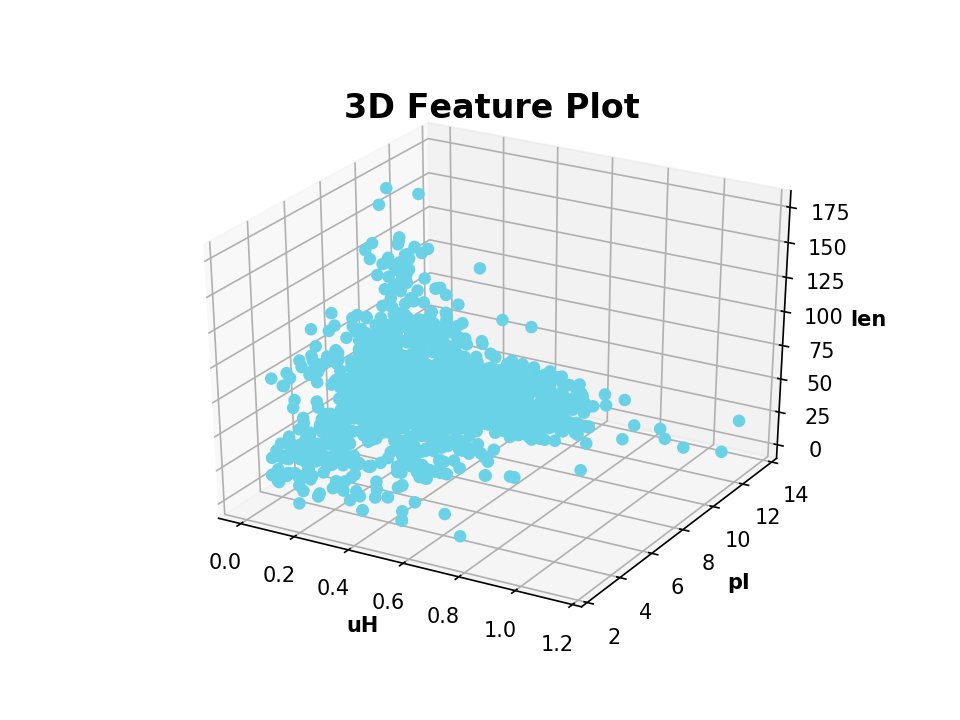
-
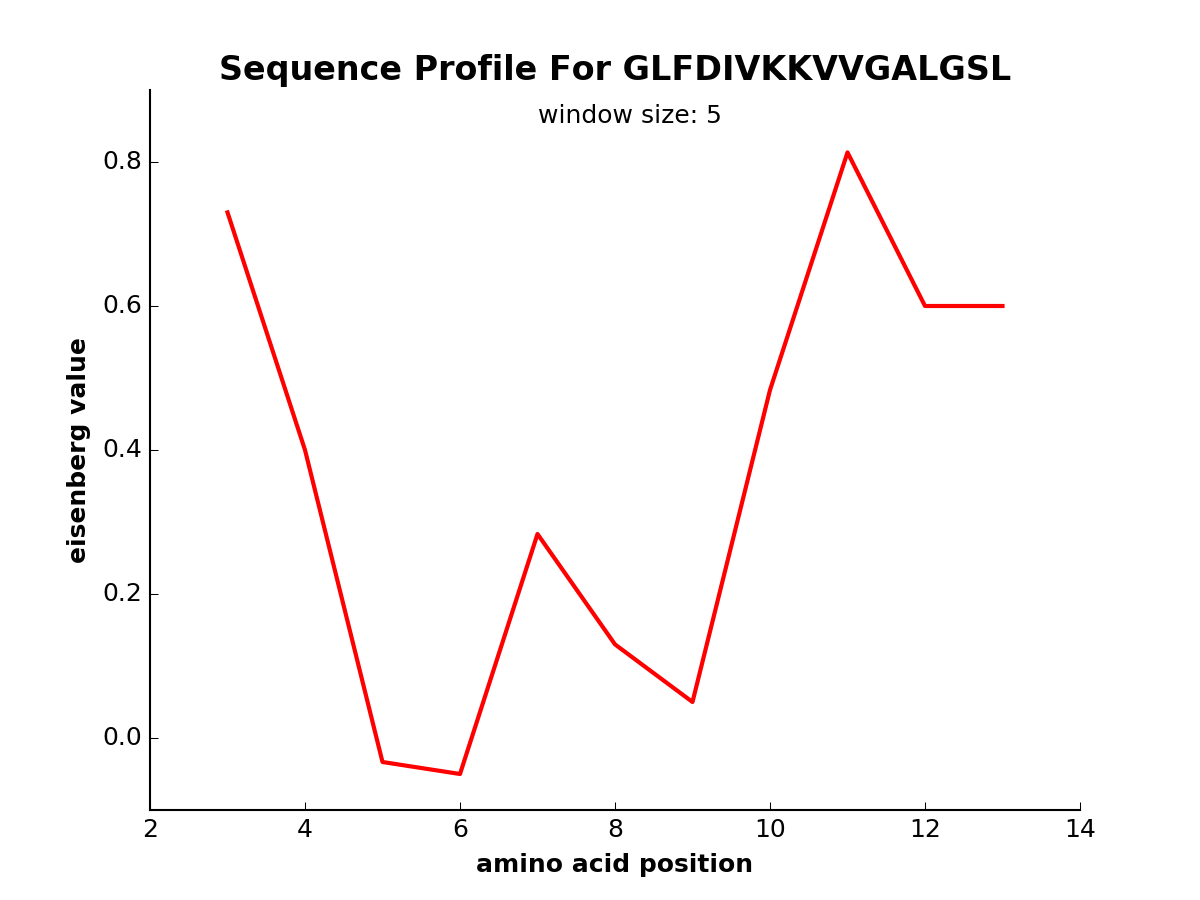
modlamp.plot.plot_profile(sequence, window=5, scalename='Eisenberg', filename=None, color='red', seq=False, ylim=None)[source]¶ Function to generate sequence profile plots of a given amino acid scale or a moment thereof.
Note
plot_profile()can only plot one-dimensional amino acid scales given inmodlamp.descriptors.PeptideDescriptor.- Parameters
sequence – {str} Peptide sequence for which the profile should be plotted.
window – {int, uneven} Window size for which the average value is plotted for the center amino acid.
scalename – {str} Amino acid scale to be used to describe the sequence.
filename – {str} Filename where to safe the plot. default = None –> show the plot
color – {str} Color of the plot line.
seq – {bool} Whether the amino acid sequence should be plotted as the title.
ylim – {tuple of float} Y-Axis limits. Provide as tuple, e.g. (0.5, -0.2)
- Returns
a profile plot of the input sequence interactively or with the specified filename
- Example
>>> plot_profile('GLFDIVKKVVGALGSL', scalename='eisenberg')
New in version v2.1.5.
-
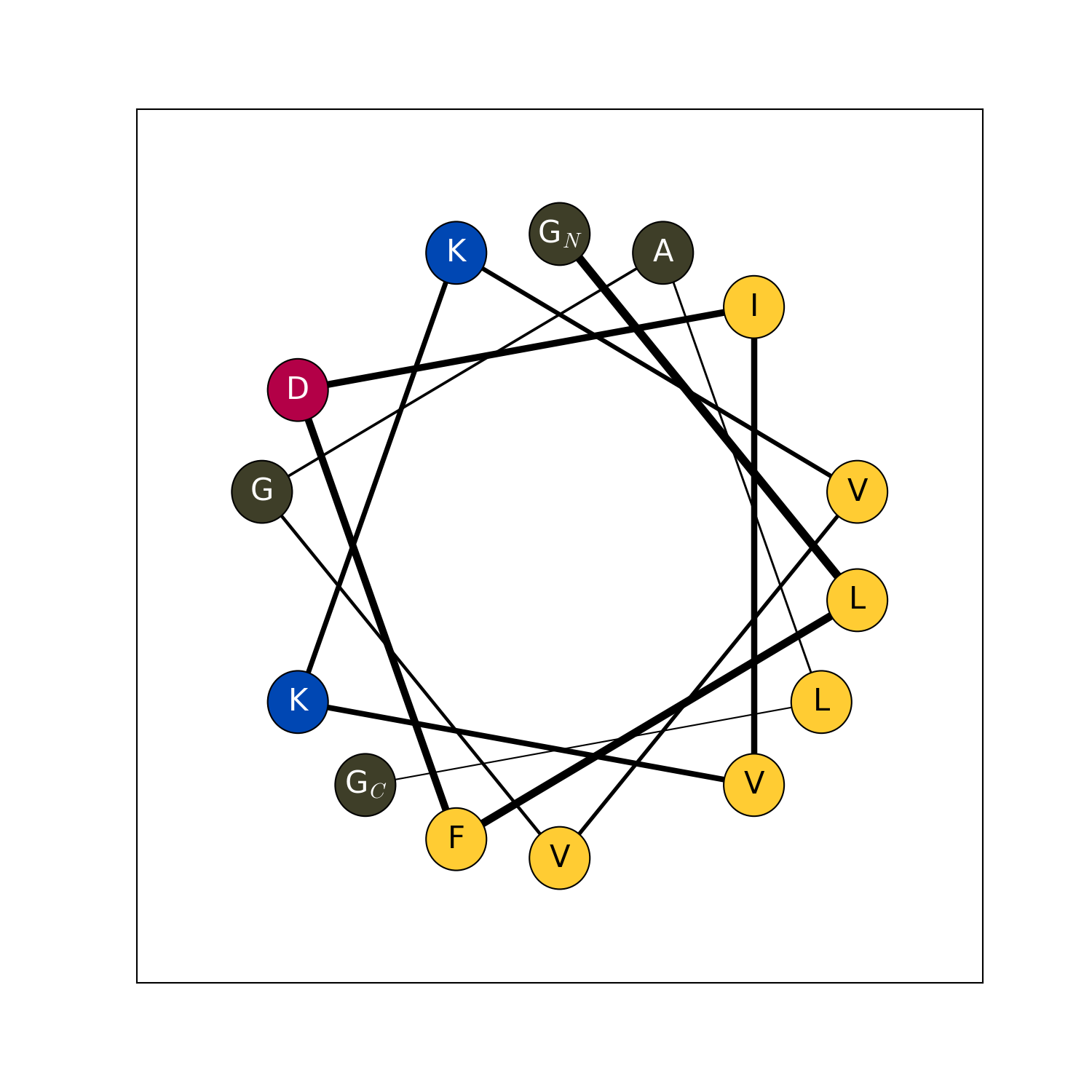
modlamp.plot.helical_wheel(sequence, colorcoding='rainbow', lineweights=True, filename=None, seq=False, moment=False)[source]¶ A function to project a given peptide sequence onto a helical wheel plot. It can be useful to illustrate the properties of alpha-helices, like positioning of charged and hydrophobic residues along the sequence.
- Parameters
sequence – {str} the peptide sequence for which the helical wheel should be drawn
colorcoding – {str} the color coding to be used, available: rainbow, charge, polar, simple, amphipathic, none
lineweights – {boolean} defines whether connection lines decrease in thickness along the sequence
filename – {str} filename where to safe the plot. default = None –> show the plot
seq – {bool} whether the amino acid sequence should be plotted as a title
moment – {bool} whether the Eisenberg hydrophobic moment should be calculated and plotted
- Returns
a helical wheel projection plot of the given sequence (interactively or in filename)
- Example
>>> helical_wheel('GLFDIVKKVVGALG') >>> helical_wheel('KLLKLLKKLLKLLK', colorcoding='charge') >>> helical_wheel('AKLWLKAGRGFGRG', colorcoding='none', lineweights=False) >>> helical_wheel('ACDEFGHIKLMNPQRSTVWY')
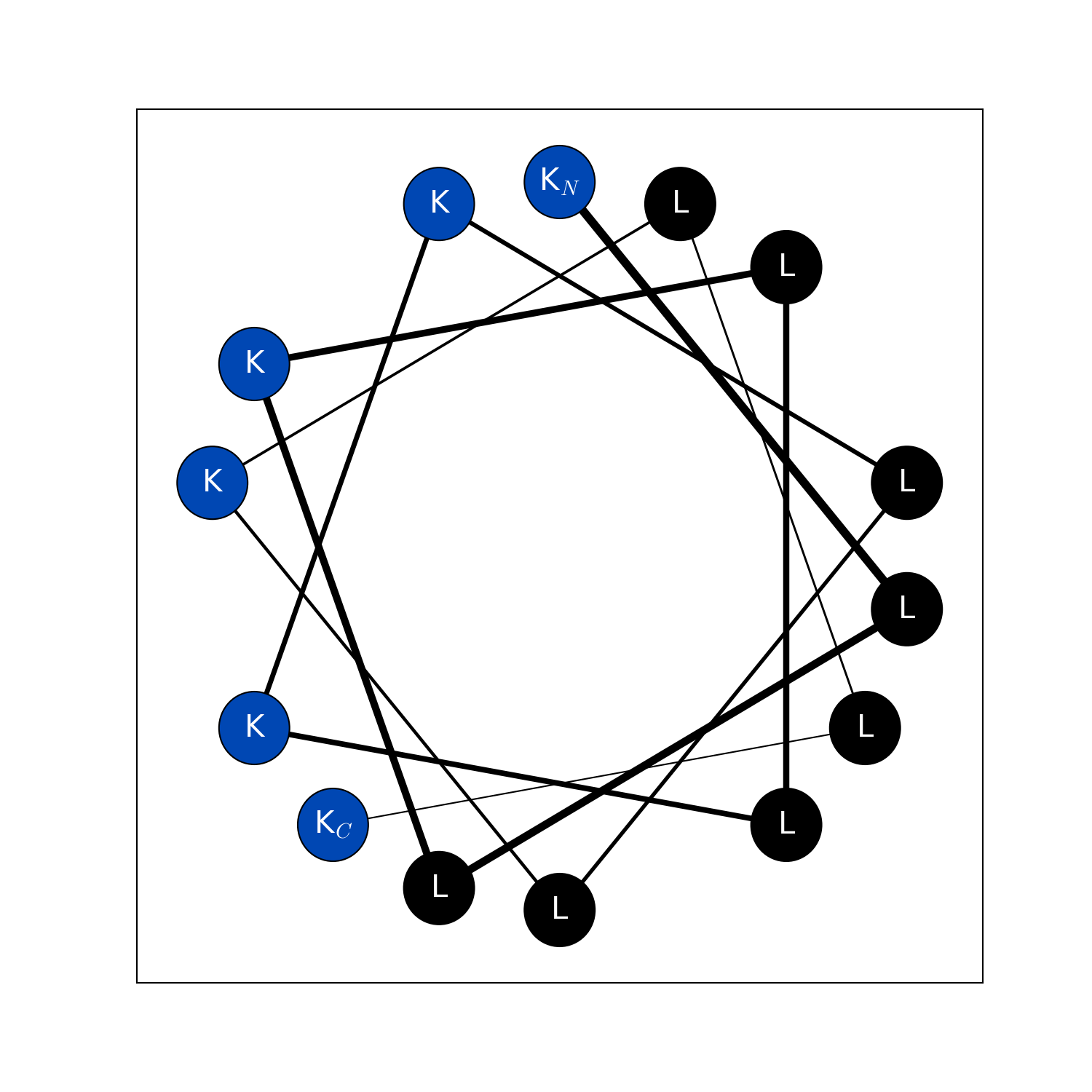
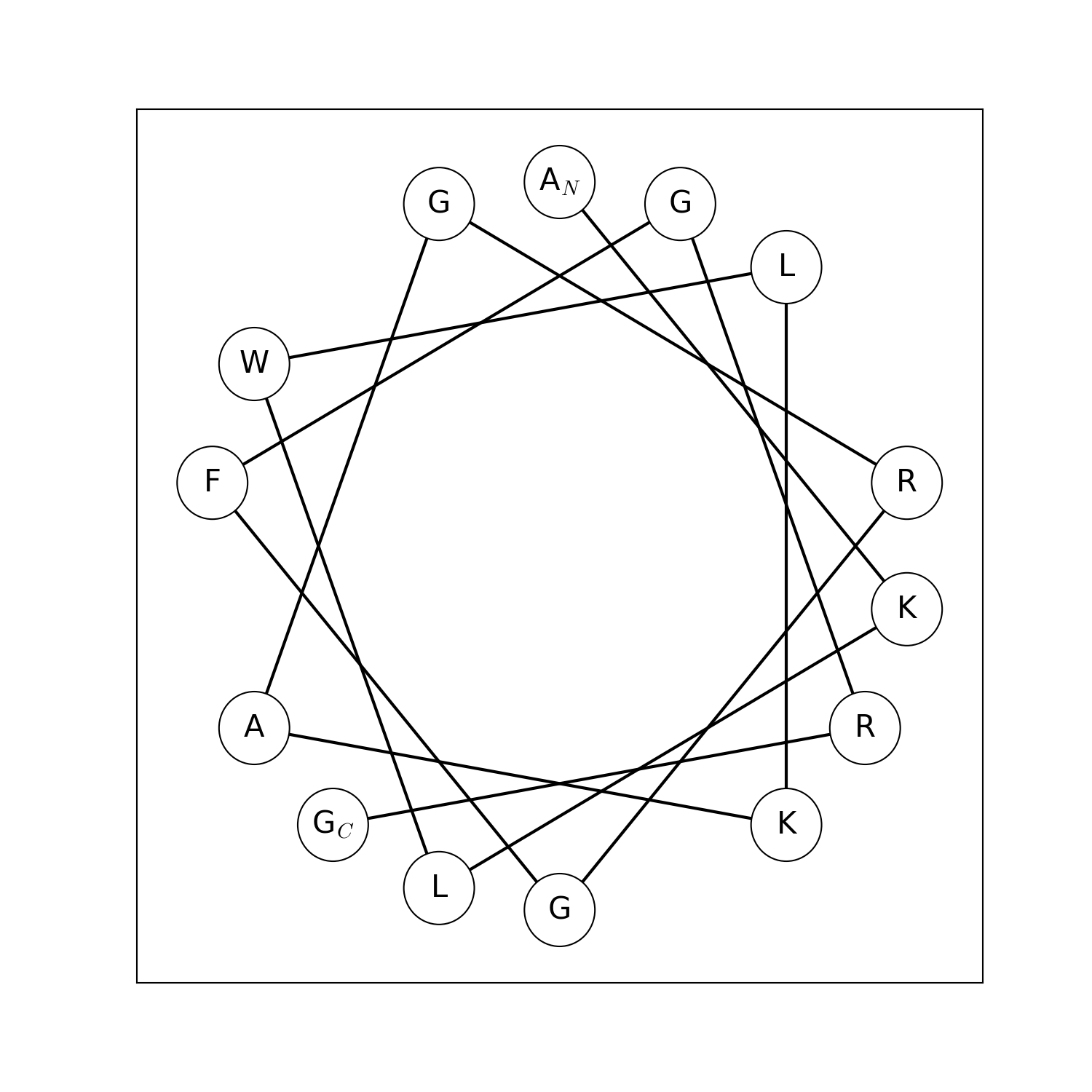
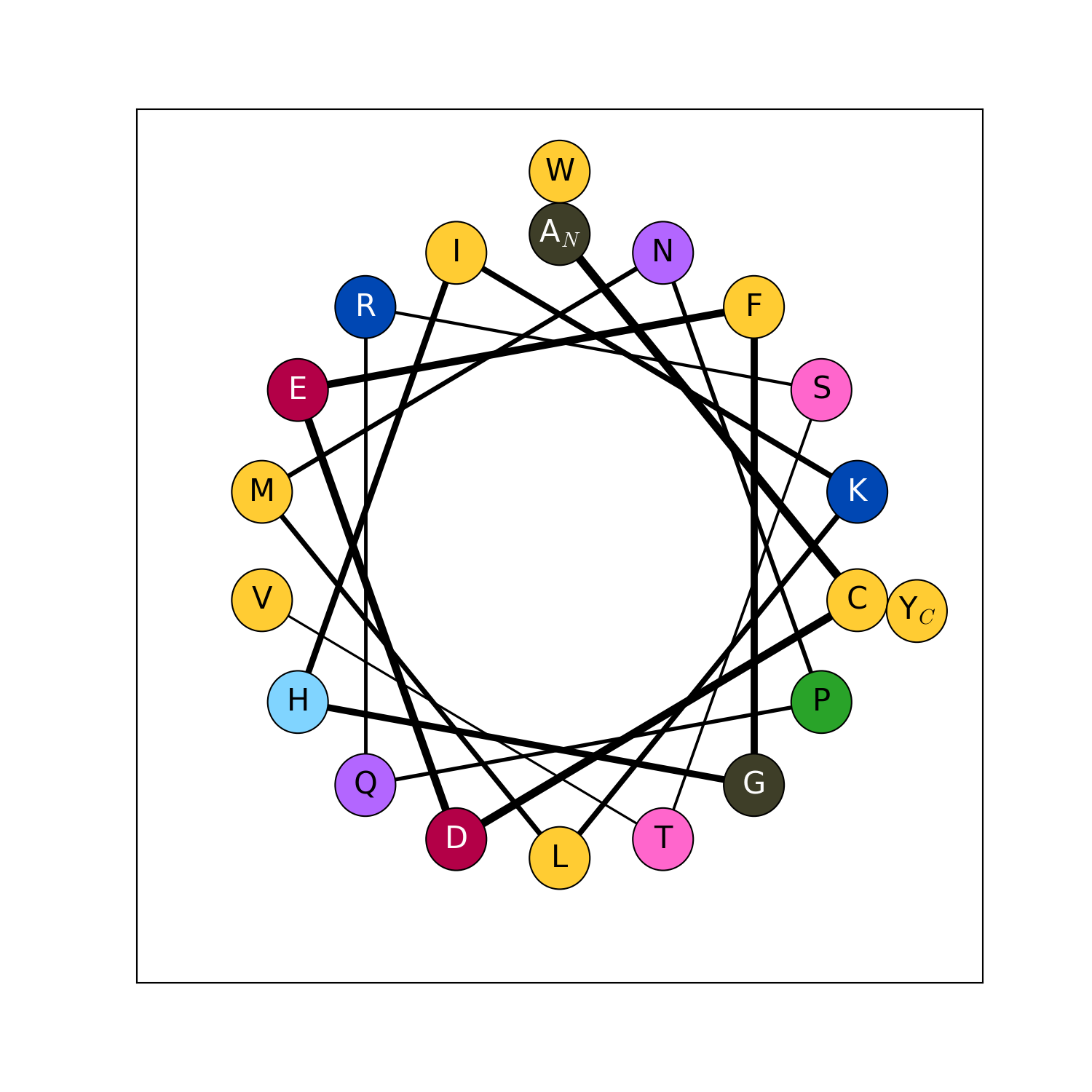
New in version v2.1.5.
-
modlamp.plot.plot_pde(data, title=None, axlabels=None, filename=None, legendloc=2, x_min=0, x_max=1, colors=None, alpha=0.2)[source]¶ A function to plot probability density estimations of given data vectors / matrices (row wise)
- Parameters
data – {list / array} data of which underlying probability density function should be estimated and plotted.
title – {str} plot title
axlabels – {list of str} list containing the axis labels for the plot
filename – {str} filename where to safe the plot. default = None –> show the plot
legendloc – {int} location of the figures legend. 1 = top right, 2 = top left …
x_min – {number} x-axis minimum
x_max – {number} x-axis maximum
colors – {list} list of colors (readable by matplotlib, e.g. hex) to be used to plot different data classes
alpha – {float} color alpha for filling pde curve
- Example
>>> data = np.random.random((3,100)) >>> plot_pde(data)
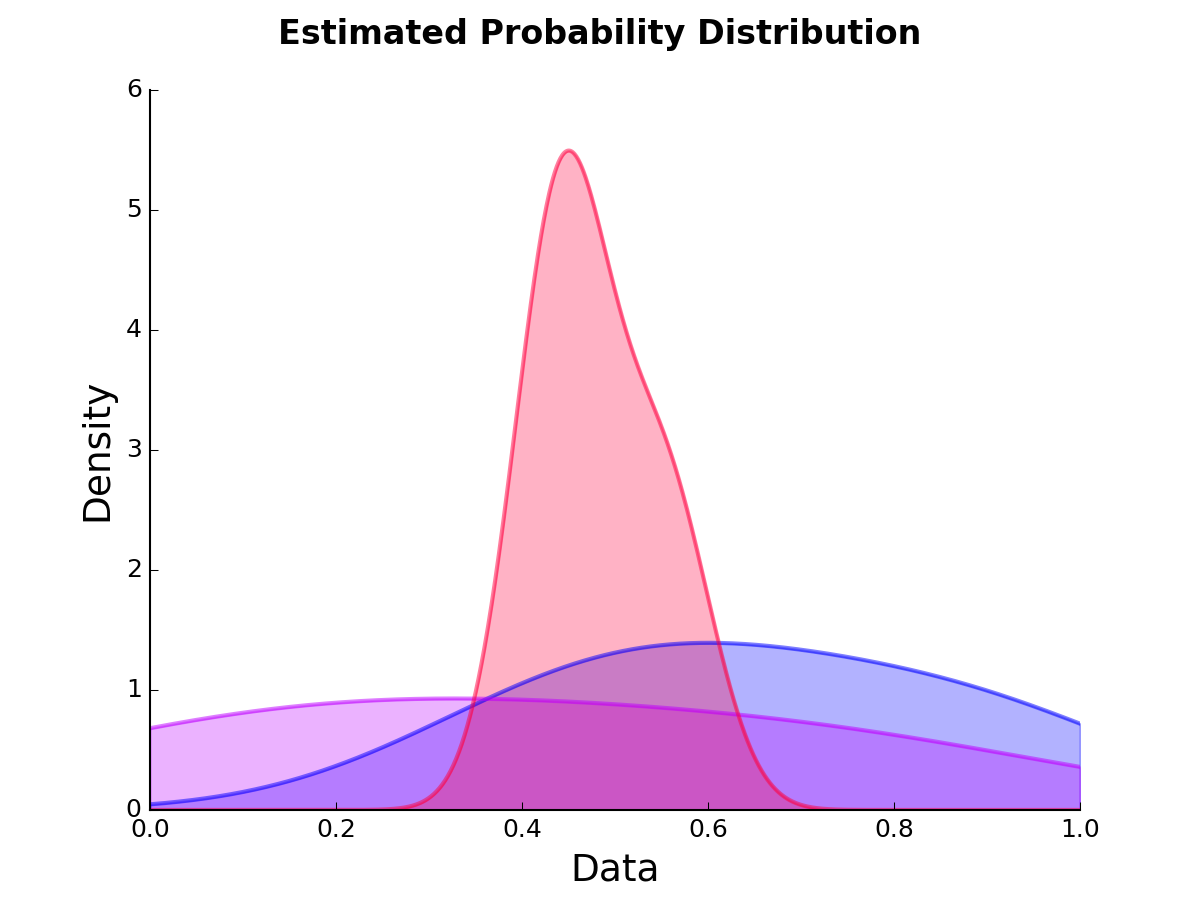
New in version v2.2.1.
-
modlamp.plot.plot_violin(x, colors=None, bp=False, filename=None, title=None, axlabels=None, y_min=0, y_max=1)[source]¶ create violin plots out of given data array (adapted from Flavio Coelho.)
- Parameters
x – {numpy.array} data to be plotted
colors – {str or list} face color of the violin plots, can also be list of colors with same dimension as x
bp – {bool} print a box blot inside violin
filename – {str} location / filename where to save the plot to. default = None –> show the plot
title – {str} Title of the plot.
axlabels – {list of str} list containing the axis labels for the plot
y_min – {number} y-axis minimum.
y_max – {number} y-axis maximum.
- Example
>>> data = np.random.normal(size=[5, 100]) >>> plot_violin(data, colors=['#0B486B', '#0B486B', '#0B486B', '#CFF09E', '#CFF09E'], bp=True, y_min=-3, y_max=3)
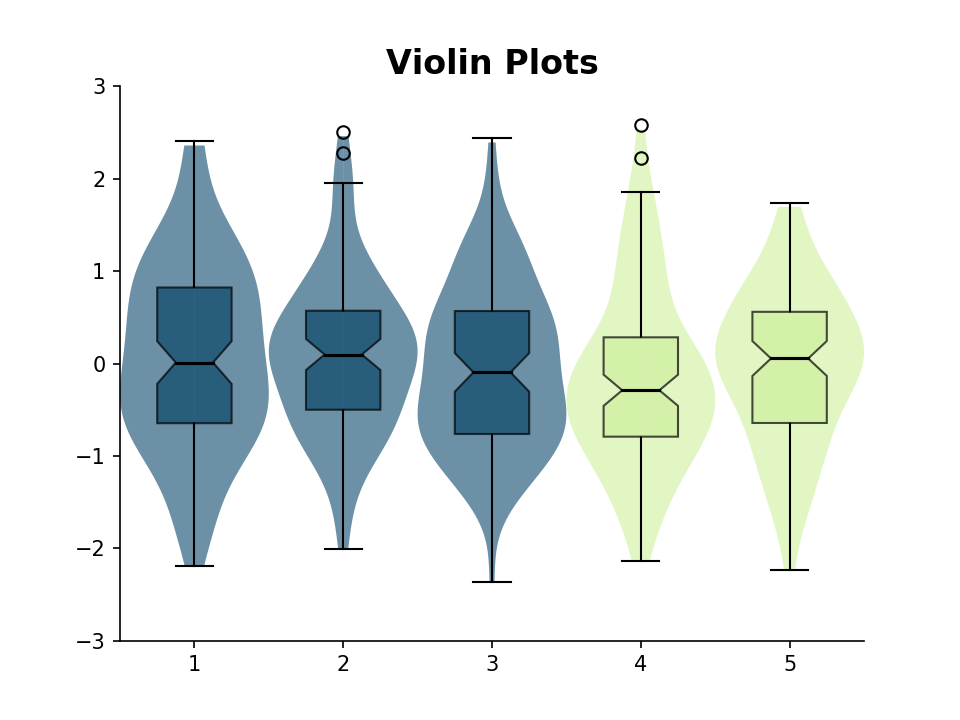
New in version v2.2.2.
-
modlamp.plot.plot_aa_distr(sequences, color='#83AF9B', filename=None)[source]¶ Method to plot the amino acid distribution of a given list of sequences
- Parameters
sequences – {list} list of sequences to calculate the amino acid distribution fore
color – {str} color to be used (matplotlib style / hex)
filename – {str} location / filename where to save the plot to. default = None –> show the plot
- Example
>>> plot_aa_distr(['KLLKLLKKLLKLLK', 'WWRRWWRAARWWRRWWRR', 'ACDEFGHKLCMNPQRSTVWY', 'GGGGGIIKLWGGGGGGGGGGGGG'])
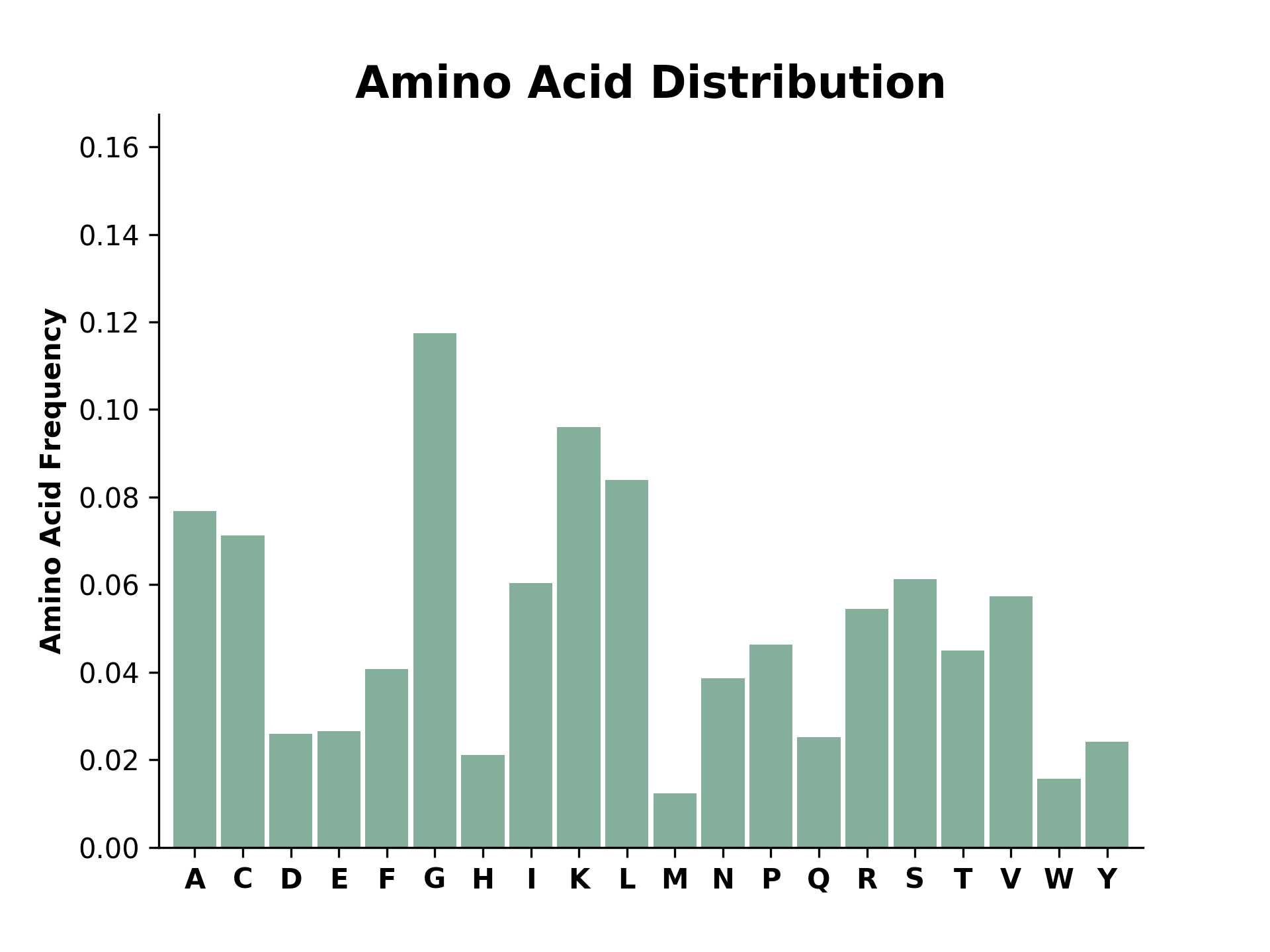
New in version v2.2.5.
2.6. modlamp.ml¶
This module contains different functions to facilitate machine learning with peptides, mainly making use of the scikit-learn Python package. Two machine learning models are available, whose parameters can be tuned. For more information on the machine learning modules please check the scikit-learn documentation.
The two available machine learning models in this module are:
Model |
Reference [1] |
|---|---|
Support Vector Machine |
|
Random Forest |
[1] F. Pedregosa et al., J. Mach. Learn. Res. 2011, 12, 2825–2830.
The following functions are included in this module:
Function name |
Description |
|---|---|
Performs a grid search on different model parameters and returns the best fitted model and its performance as the cross-validation MCC. |
|
Plotting a validation curve for any parameter from the grid search. |
|
Predict the class labels or class probabilities for given peptides. |
|
Evaluate the performance of a given model through cross-validation. |
|
Evaluate the performance of a given model through test-set prediction. |
New in version 2.2.0.
-
modlamp.ml.train_best_model(model, x_train, y_train, sample_weights=None, scaler=StandardScaler(copy=True, with_mean=True, with_std=True), score=make_scorer(matthews_corrcoef), param_grid=None, n_jobs=-1, cv=10)[source]¶ This function performs a parameter grid search on a selected classifier model and peptide training data set. It returns a scikit-learn pipeline that performs standard scaling and contains the best model found by the grid search according to the Matthews correlation coefficient. (see sklearn.preprocessing, sklearn.grid_search, sklearn.pipeline.Pipeline).
- Parameters
model – {str} model to train. Choose between
'svm'(Support Vector Machine) or'rf'(Random Forest).x_train – {array} descriptor values for training data.
y_train – {array} class values for training data.
sample_weights – {array} sample weights for training data.
scaler – {scaler} scaler to use in the pipe to scale data prior to training. Choose from
sklearn.preprocessing, e.g.StandardScaler(),MinMaxScaler(),Normalizer().score – {metrics instance} scoring function built from make_scorer() or a predefined value in string form (choose from the scikit-learn scoring-parameters).
param_grid –
{dict} parameter grid for the gridsearch (see sklearn.grid_search).
n_jobs – {int} number of parallel jobs to use for calculation. if
-1, all available cores are used.cv – {int} number of folds for cross-validation.
- Returns
best estimator pipeline.
Default parameter grids:
Model
Parameter grid
SVM Model
param_grid = [{‘clf__C’: param_range, ‘clf__kernel’: [‘linear’]}, {‘clf__C’: param_range, ‘clf__gamma’: param_range, ‘clf__kernel’: [‘rbf’]}]
Random Forest
- param_grid = [{‘clf__n_estimators’: [10, 100, 500],
‘clf__max_features’: [‘sqrt’, ‘log2’], ‘clf__bootstrap’: [True], ‘clf__criterion’: [“gini”]}]
Useful methods implemented in scikit-learn:
Method
Description
fit(x, y)
fit the model with the same parameters to new training data.
score(x, y)
get the score of the model for test data.
predict(x)
get predictions for new data.
predict_proba(x)
get probability predictions for [class0, class1]
get_params()
get parameters of the trained model
- Example
>>> from modlamp.ml import train_best_model >>> from modlamp.datasets import load_ACPvsRandom >>> from modlamp.descriptors import PeptideDescriptor
Loading a dataset for training:
>>> data = load_ACPvsRandom() >>> len(data.sequences) 826 >>>list(data.target_names) ['Random', 'ACP']
Calculating the pepCATS descriptor values in auto-correlation modality:
>>> descr = PeptideDescriptor(data.sequences, scalename='pepcats') >>> descr.calculate_autocorr(7) >>> descr.descriptor.shape (826, 42) >>> descr.descriptor array([[ 1. , 0.15 , 0. , ..., 0.35714286, 0.21428571, 0. ], [ 0.64 , 0.12 , 0.32 , ..., 0.05263158, 0. , 0. ], [ 1. , 0.23809524, 0. , ..., 0.53333333, 0.26666667, 0. ], ..., [ 0.5 , 0.22222222, 0.44444444, ..., 0.33333333, 0. , 0. ], [ 0.70588235, 0.17647059, 0.23529412, ..., 0.09090909, 0.09090909, 0. ], [ 0.6875 , 0.1875 , 0.1875 , ..., 0.2 , 0. , 0. ]])
Training an SVM model on this descriptor data:
>>> X_train = descr.descriptor >>> y_train = data.target >>> best_svm_model = train_best_model('svm', X_train, y_train) Best score (scorer: make_scorer(matthews_corrcoef)) and parameters from a 10-fold cross validation: 0.739995453978 {'clf__gamma': 0.1, 'clf__C': 10.0, 'clf__kernel': 'rbf'}
>>> best_svm_model.get_params() {'clf': SVC(C=10.0, cache_size=200, class_weight='balanced', coef0=0.0, decision_function_shape=None, degree=3, gamma=0.1, kernel='rbf', max_iter=-1, probability=True, random_state=1, shrinking=True, tol=0.001, verbose=False), ... 'steps': [('scl', StandardScaler(copy=True, with_mean=True, with_std=True)), ('clf', SVC(C=10.0, cache_size=200, class_weight='balanced', coef0=0.0, decision_function_shape=None, degree=3, gamma=0.1, kernel='rbf', max_iter=-1, probability=True, random_state=1, shrinking=True, tol=0.001, verbose=False))]}
-
modlamp.ml.plot_validation_curve(classifier, x_train, y_train, param_name, param_range, cv=10, score=make_scorer(matthews_corrcoef), title='Validation Curve', xlab='parameter range', ylab='MCC', n_jobs=-1, filename=None)[source]¶ This function plots a cross-validation curve for the specified classifier on all tested parameters given in the option
param_range.- Parameters
classifier – {classifier instance} classifier or validation curve (e.g. sklearn.svm.SVC).
x_train – {array} descriptor values for training data.
y_train – {array} class values for training data.
param_name – {string} parameter to assess in the validation curve plot. For SVM, “clf__C” (C parameter), “clf__gamma” (gamma parameter). For Random Forest, “clf__n_estimators” (number of trees) “clf__max_depth” (max num of branches per tree, “clf__min_samples_split” (min number of samples required to split an internal tree node), “clf__min_samples_leaf” (min number of samples in newly created leaf).
param_range – {list} parameter range for the validation curve.
cv – {int} number of folds for cross-validation.
score – {metrics instance} scoring function built from make_scorer() or a predefined value in string form sklearn.model_evaluation.scoring-parameter.
title – {str} graph title
xlab – {str} x axis label.
ylab – {str} y axis label.
n_jobs – {int} number of parallel jobs to use for calculation. if
-1, all available cores are used.filename – {str} if filename given the figure is stored in the specified path.
- Returns
plot of the validation curve.
- Example
>>> from modlamp.ml import train_best_model >>> from modlamp.datasets import load_ACPvsRandom >>> from modlamp.descriptors import PeptideDescriptor
Loading a dataset for training:
>>> data = load_ACPvsRandom() >>> len(data.sequences) 826 >>>list(data.target_names) ['Random', 'ACP']
Calculating the pepCATS descriptor values in auto-correlation modality:
>>> descr = PeptideDescriptor(data.sequences, scalename='pepcats') >>> descr.calculate_autocorr(7) >>> descr.descriptor.shape (826, 42) >>> descr.descriptor array([[ 1. , 0.15 , 0. , ..., 0.35714286, 0.21428571, 0. ], [ 0.64 , 0.12 , 0.32 , ..., 0.05263158, 0. , 0. ], [ 1. , 0.23809524, 0. , ..., 0.53333333, 0.26666667, 0. ], ..., [ 0.5 , 0.22222222, 0.44444444, ..., 0.33333333, 0. , 0. ], [ 0.70588235, 0.17647059, 0.23529412, ..., 0.09090909, 0.09090909, 0. ], [ 0.6875 , 0.1875 , 0.1875 , ..., 0.2 , 0. , 0. ]])
Training an SVM model with this data:
>>> X_train = descr.descriptor >>> y_train = data.target >>> best_svm_model = train_best_model('svm', X_train, y_train) Best score (scorer: make_scorer(matthews_corrcoef)) and parameters from a 10-fold cross validation: 0.739995453978 {'clf__gamma': 0.1, 'clf__C': 10.0, 'clf__kernel': 'rbf'}
>>> plot_validation_curve(best_svm_model, X_train, y_train, param_name='clf__gamma', param_range=[0.0001, 0.001, 0.01, 0.1, 1, 10, 100, 1000])
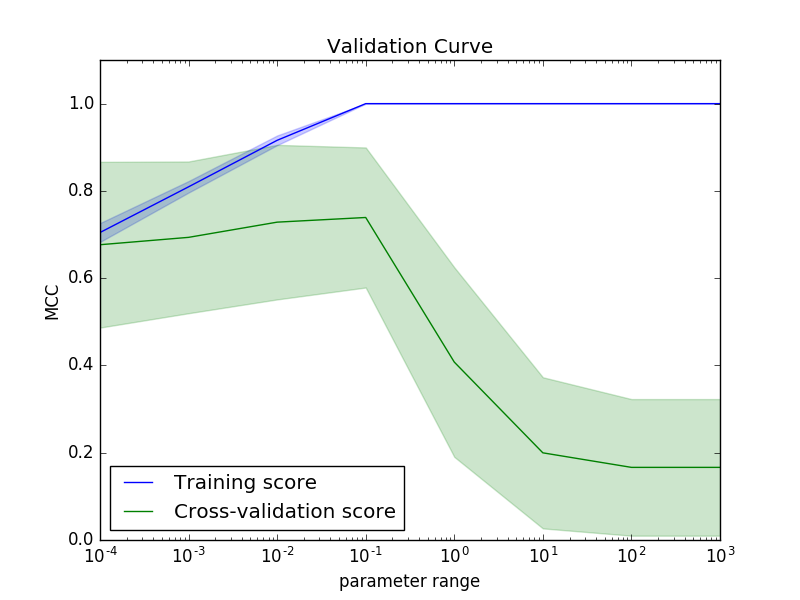
-
modlamp.ml.predict(classifier, x, seqs, names=None, y=None, filename=None)[source]¶ This function can be used to predict novel peptides with a trained classifier model. The function returns a
pandas.DataFramewith predictions using the specified estimator and test data. If true class is provided, it returns the scoring value for the test data.- Parameters
classifier – {classifier instance} classifier used for predictions.
x – {array} descriptor values of the peptides to be predicted.
seqs – {list} sequences of the peptides in
x.names – {list} (optional) names of the peptides in
x.y – {array} (optional) true (known) classes of the peptides.
filename – {string} (optional) output filename to store the predictions to (
.csvformat); ifNone: not saved.
- Returns
pandas.DataFramecontaining predictions forx.P_class0andP_class1are the predicted probability of the peptide belonging to class 0 and class 1, respectively.- Example
>>> from modlamp.ml import train_best_model, predict >>> from modlamp.datasets import load_ACPvsRandom >>> from modlamp.descriptors import PeptideDescriptor >>> from modlamp.sequences import Helices
Loading data for model training:
>>> data = load_ACPvsRandom()
Calculating descriptor values from the data:
>>> desc = PeptideDescriptor(data.sequences, scalename='pepcats') >>> desc.calculate_autocorr(7) >>> best_svm_model = train_best_model('svm', desc.descriptor, data.target)
Generating 10 de novo helical sequences to predict their activity:
>>> H = Helices(seqnum=10, lenmin=7, lenmax=30) >>> H.generate_sequences()
Calculating descriptor values for the newly generated sequences:
>>> descH = PeptideDescriptor(H.sequences, scalename='pepcats') >>> descH.calculate_autocorr(7)
>>> df = predict(best_svm_model, x=descH.descriptor, seqs=descH.sequences) >>> df.head(3) # all three shown sequences are predicted active (class 1) Sequence P_class0 P_class1 IAGKLAKVGLKIGKIGGKLVKGVLK 0.009167 0.990833 LGVRVLRIIIR 0.007239 0.992761 VGIRLARGVGRIG 0.071436 0.928564
-
modlamp.ml.score_cv(classifier, x, y, sample_weights=None, cv=10, shuffle=True)[source]¶ This function can be used to evaluate the performance of selected classifier model. It returns the average cross-validation scores for the specified scoring metrics in a
pandas.DataFrame.- Parameters
classifier – {classifier instance} a classifier model to be evaluated.
x – {array} descriptor values for training data.
y – {array} class values for training data.
sample_weights – {array} weights for training data.
cv – {int} number of folds for cross-validation.
shuffle – {bool} suffle data before making the K-fold split.
- Returns
pandas.DataFramecontaining the cross validation scores for the specified metrics.- Example
>>> from modlamp.ml import train_best_model, score_cv >>> from modlamp.datasets import load_ACPvsRandom >>> from modlamp.descriptors import PeptideDescriptor
Loading data for model training:
>>> data = load_ACPvsRandom()
Calculating descriptor values from the data:
>>> desc = PeptideDescriptor(data.sequences, scalename='pepcats') >>> desc.calculate_autocorr(7) >>> best_svm_model = train_best_model('svm', desc.descriptor, data.target)
Get the cross-validation scores:
>>> score_cv(best_svm_model, desc.descriptor, data.target, cv=5) CV_0 CV_1 CV_2 CV_3 CV_4 mean std MCC 0.785 0.904 0.788 0.757 0.735 0.794 0.059 accuracy 0.892 0.952 0.892 0.880 0.867 0.896 0.029 precision 0.927 0.974 0.953 0.842 0.884 0.916 0.048 recall 0.864 0.925 0.854 0.889 0.864 0.879 0.026 f1 0.894 0.949 0.901 0.865 0.874 0.896 0.029 roc_auc 0.893 0.951 0.899 0.881 0.868 0.898 0.028
-
modlamp.ml.score_testset(classifier, x_test, y_test, sample_weights=None)[source]¶ Returns the test set scores for the specified scoring metrics in a
pandas.DataFrame. The calculated metrics are Matthews correlation coefficient, accuracy, precision, recall, f1 and area under the Receiver-Operator Curve (roc_auc). See sklearn.metrics for more information.- Parameters
classifier – {classifier instance} pre-trained classifier used for predictions.
x_test – {array} descriptor values of the test data.
y_test – {array} true class values of the test data.
sample_weights – {array} weights for the test data.
- Returns
pandas.DataFramecontaining the cross validation scores for the specified metrics.- Example
>>> from modlamp.ml import train_best_model, score_testset >>> from sklearn.model_selection import train_test_split >>> from modlamp.datasets import load_ACPvsRandom >>> from modlamp import descriptors >>> data = load_ACPvsRandom()
Calculating descriptor values from the data
>>> desc = descriptors.PeptideDescriptor(data.sequences, scalename='pepcats') >>> desc.calculate_autocorr(7)
Splitting data into train and test sets
>>> X_train, X_test, y_train, y_test = train_test_split(desc.descriptor, data.target, test_size = 0.33)
Training a SVM model on the training set
>>> best_svm_model = train_best_model('svm', X_train,y_train)
Calculating the scores of the predictions on the test set
>>> score_testset(best_svm_model, X_test, y_test) Metrics Scores MCC 0.839 accuracy 0.920 precision 0.924 recall 0.910 f1 0.917 roc_auc 0.919
2.7. modlamp.wetlab¶
This module incorporates functions to load raw data files from wetlab experiments, calculate different characteristics and plot.
Class |
Data |
|---|---|
Class for handling Circular Dichroism data. |
-
class
modlamp.wetlab.CD(directory, wmin, wmax, amide=True, pathlen=1)[source]¶ Class to handle circular dichroism data files and calculate several ellipticity and helicity values. The class can handle data files of the Applied Photophysics type.
For explanations of different units used in CD spectroscopy, visit https://www.photophysics.com/resources/7-cd-units-conversions and read the following publication:
Greenfield, Nat. Protoc. 2006, 1, 2876–2890.
Note
All files which should be read must have 4 header lines as shown in the image below. CD data to be read must start in line 5 (separated in 2 columns: Wavelength and Signal).
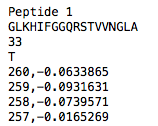
First line: Molecule Name
Second line: Sequence
Third line: concentration in µM
Fourth line: solvent
Recognized solvents are W for water and T for TFE.
-
__init__(directory, wmin, wmax, amide=True, pathlen=1)[source]¶ Init method for class CD.
- Parameters
directory – {str} directory containing all data files to be read. Files need a .csv ending
wmin – {int} smalles wavelength measured
wmax – {int} highest wavelength measured
amide – {bool} specifies whether the sequences have amidated C-termini
pathlen – {float} cuvette path length in mm
- Example
>>> cd = CD('/path/to/your/folder', 185, 260) >>> cd.filenames ['160819_Pep1_T_smooth.csv', '160819_Pep1_W_smooth.csv', '160819_Pep5_T_smooth.csv', ...] >>> cd.names ['Pep 10', 'Pep 10', 'Pep 11', 'Pep 11', ... ] >>> cd.conc_umol [33.0, 33.0, 33.0, 33.0, 33.0, 33.0, 33.0, 33.0, 33.0, 33.0, 33.0, ... ] >>> cd.meanres_mw [114.29920769230768, 114.29920769230768, 111.68257692307689, 111.68257692307689, ... ]
-
calc_molar_ellipticity()[source]¶ Method to calculate the molar ellipticity for all loaded data in
circular_dichroismaccording to the following formula:[\Theta] = (\Theta * MW) / (c * l)
- Returns
{numpy array} molar ellipticity in
molar_ellipticity- Example
>>> cd.calc_molar_ellipticity() >>> cd.molar_ellipticity array([[ 260., -1.40893636e+04], [ 259., -1.00558182e+04], [ 258., -1.25173636e+04], ...
-
calc_meanres_ellipticity()[source]¶ Method to calculate the mean residue ellipticity for all loaded data in
circular_dichroismaccording to the following formula:(\Theta * MRW) / (c * l) = [\Theta] MRW = MW / (n - 1)
With MRW = mean residue weight (g/mol), n = number of residues in the peptide, c = concentration (mg/mL) and l = cuvette path length (mm).
- Returns
{numpy array} molar ellipticity in
meanres_ellipticity- Example
>>> cd.calc_meanres_ellipticity() >>> cd.meanres_ellipticity array([[ 260. , -2669.5804196], [ 259. , -3381.3286713], [ 258. , -3872.5174825], ...
-
plot(data='mean residue ellipticity', combine='solvent', ylim=None)[source]¶ Method to generate CD plots for all read data in the initial directory.
- Parameters
data – {str} which data should be plotted (
mean residue ellipticity,molar ellipticityorcircular dichroism)combine – {str} if
solvent, overlays of different solvents will be created for the same molecule. The amino acid sequence in the header is used to find corresponding data. ifall, all data is combined in one single plot. To ignore combination, pass an empty string.ylim – {tuple} If not none, this tuple of values is taken as the minimum and maximum of the y axis
- Returns
.pdf plots saved to the directory containing the read files.
- Example
>>> cd = CD('/path/to/your/folder', 185, 260) >>> cd.calc_meanres_ellipticity() >>> cd.plot(data='mean residue ellipticity', combine='solvent')
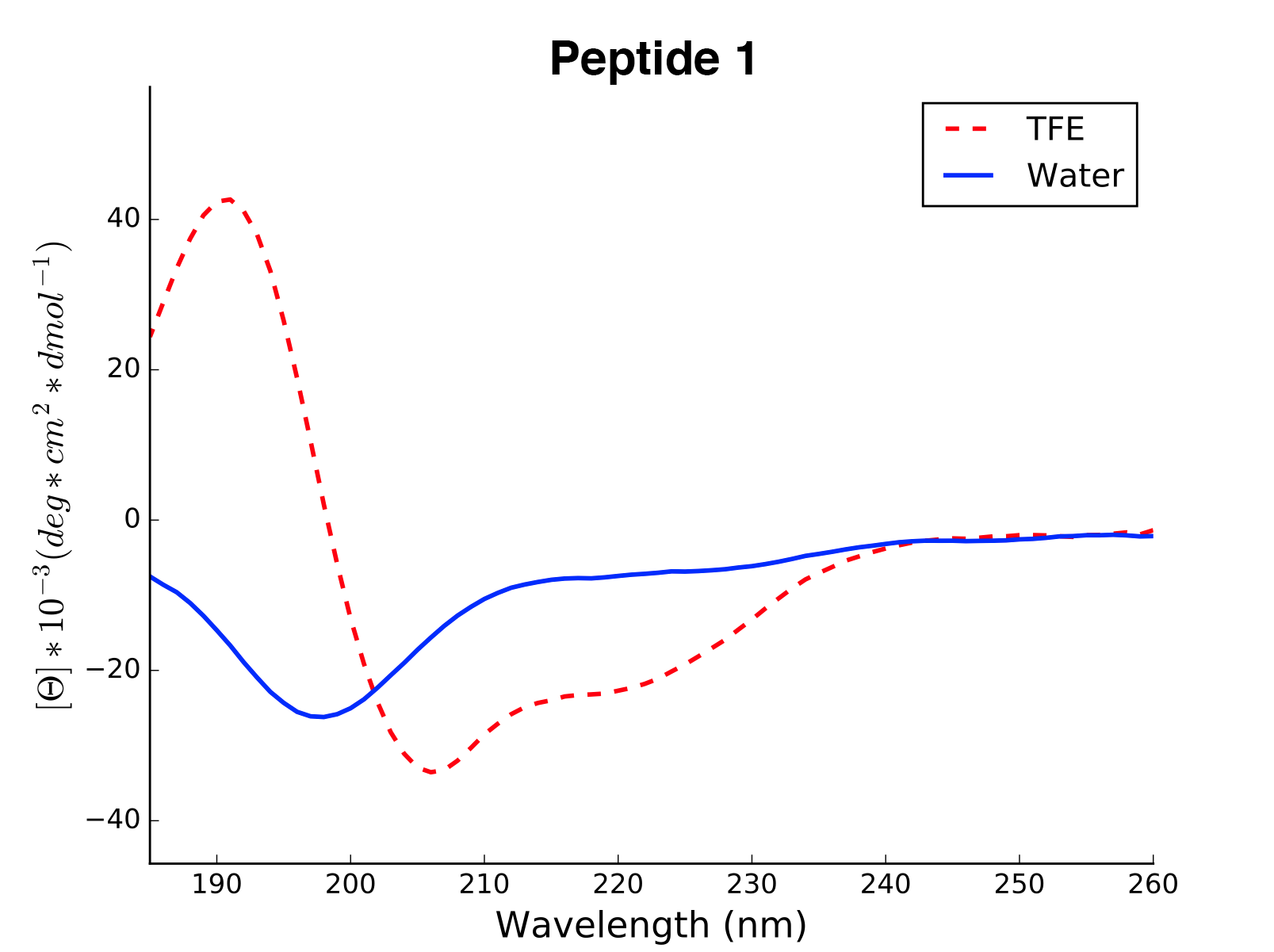
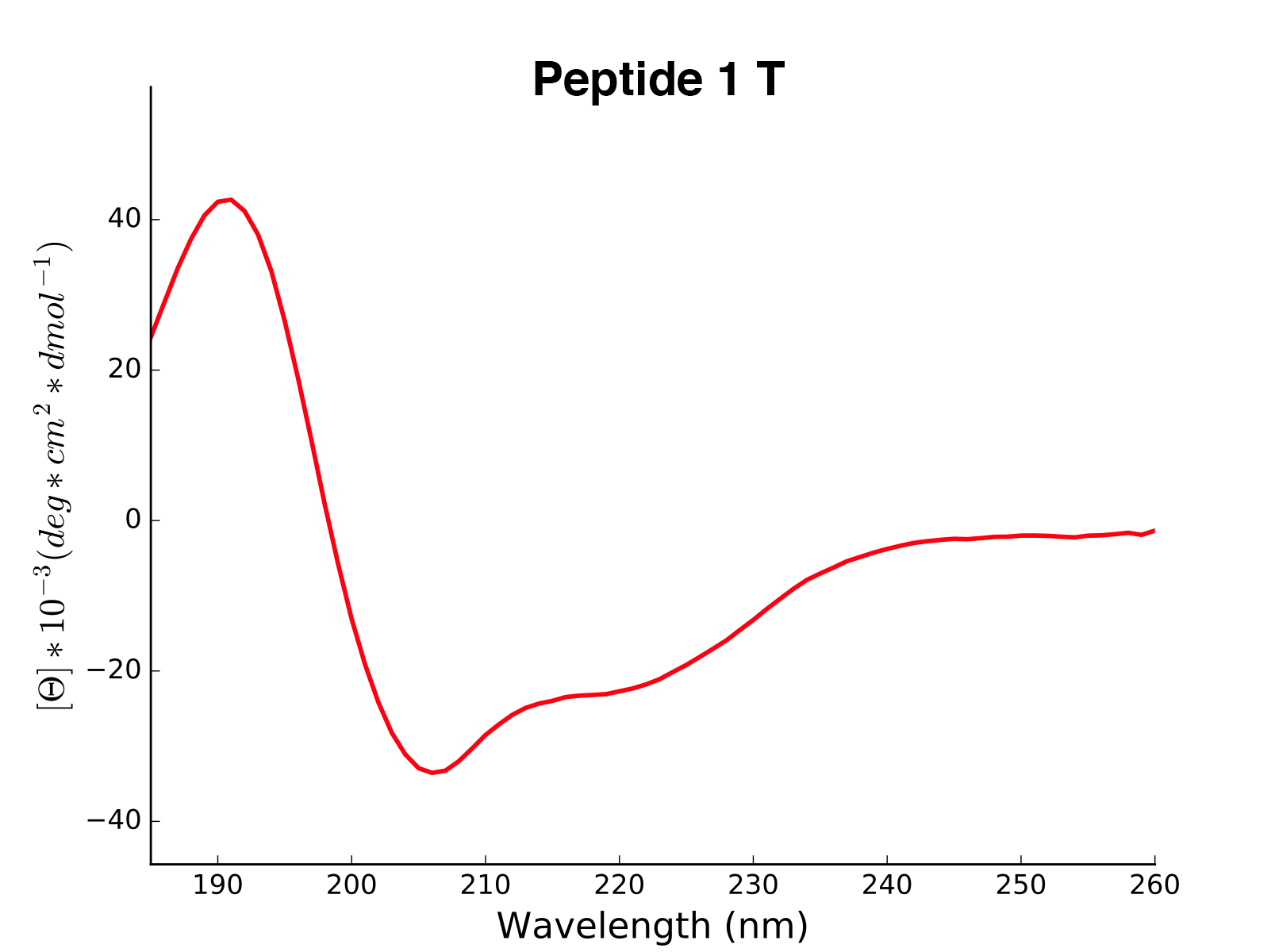
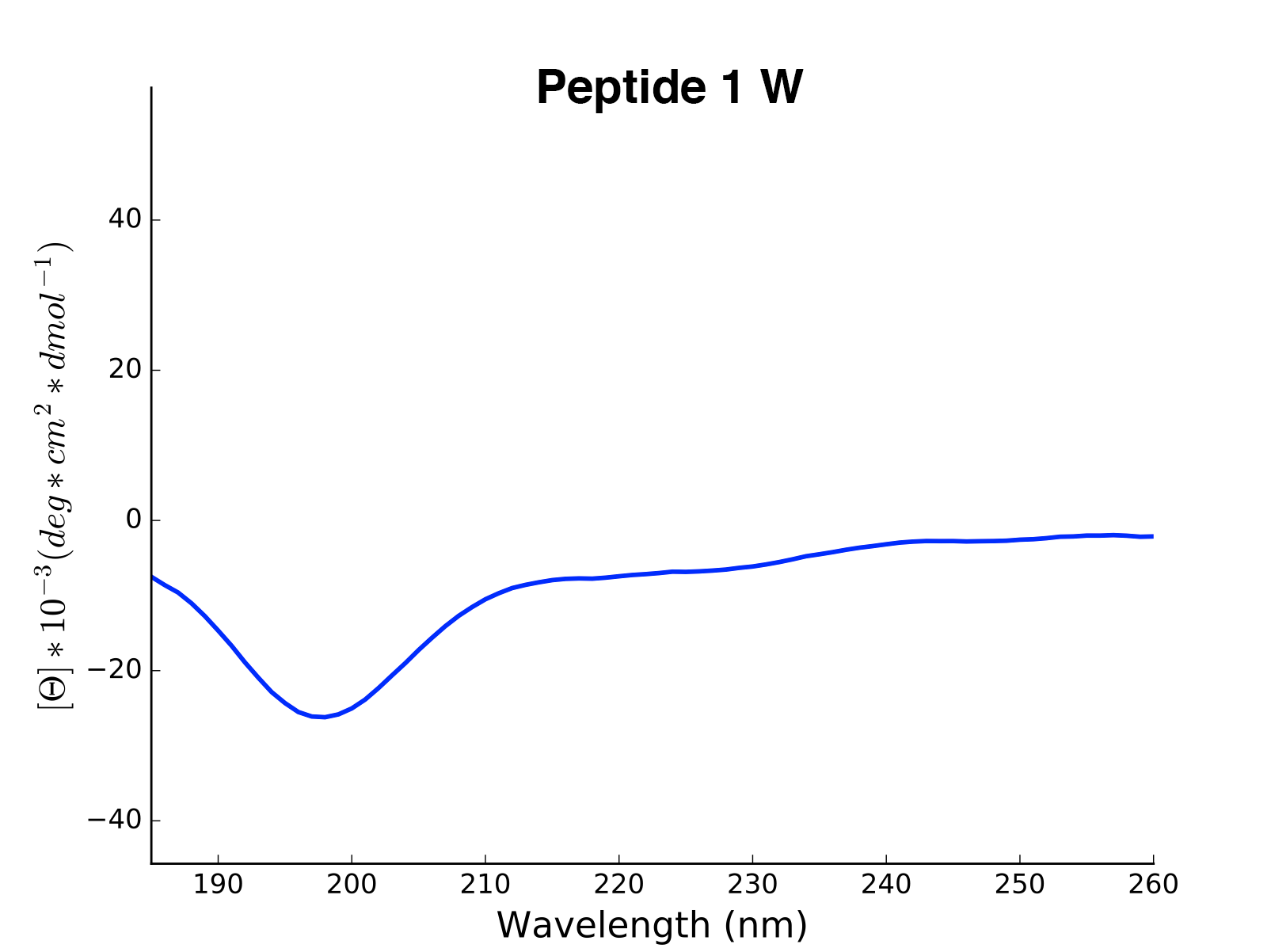
-
dichroweb(data='mean residue ellipticity')[source]¶ Method to save the calculated CD data into DichroWeb readable format (semi-colon separated). The produced files can then directly be uploaded to the DichroWeb analysis tool.
- Parameters
data – {str} which data should be plotted (
mean residue ellipticity,molar ellipticityorcircular dichroism)- Returns
.csv data files saved to the directory containing the read files.
-
helicity(temperature=24.0, k=3.5, induction=True, filename=None)[source]¶ Method to calculate the percentage of helicity out of the mean residue ellipticity data. The calculation is based on the fromula by Fairlie and co-workers:
[\Theta]_{222\infty} = (-44000 * 250 * T) * (1 - k / N)
The helicity is then calculated as the ratio of
([\Theta]_{222} / [\Theta]_{222\infty}) * 100 \%
- Reference
- Parameters
temperature – {float} experiment temperature in °C
k – {float, 2.4 - 4.5} finite length correction factor. Can be adapted to the helicity of a known peptide.
induction – {bool} wether the helical induction upon changing from one solvent to another should be calculated.
filename – {str} if given, helicity data is saved to the file “filename”.csv
- Returns
approximate helicity for every sequence in the attribute
helicity_values.- Example
>>> cd.calc_meanres_ellipticity() >>> cd.helicity(temperature=24., k=3.492185008, induction=True) >>> cd.helicity_values Name Solvent Helicity Induction 0 Aurein2.2d2 T 100.0 3.823 1 Aurein2.2d2 W 26.16 0.000 2 Klak14 T 76.38 3.048 3 Klak14 W 25.06 0.000
2.8. modlamp.analysis¶
This module can be used for diverse analysis of given peptide libraries.
-
class
modlamp.analysis.GlobalAnalysis(library, names=None)[source]¶ Base class for amino acid sequence library analysis
New in version 2.6.0.
-
__init__(library, names=None)[source]¶ - Parameters
library – {list, numpy.ndarray, pandas.DataFrame} sequence library, if 2D, the rows are considered as sub-libraries.
names – {list} list of library names to plot as labels and legend
- Example
>>> g = GlobalAnalysis(['GLFDIVKKVVGALG', 'KLLKLLKKLLKLLK', ...], names=['Library1'])
-
calc_aa_freq(plot=True, color='#83AF9B', filename=None)[source]¶ Method to get the frequency of every amino acid in the library. If the library consists of sub-libraries, the frequencies of these are calculated independently.
- Parameters
plot – {bool} whether the amino acid frequencies should be plotted in a histogram.
color – {str} color of the plot
filename – {str} filename to save the plot to, if None, the plot is shown
- Returns
{numpy.ndarray} amino acid frequencies in the attribute
aafreq. The values are oredered alphabetically.- Example
>>> g = GlobalAnalysis(sequences) # sequences being a list / array of amino acid sequences >>> g.calc_aa_freq() >>> g.aafreq array([[ 0.08250071, 0. , 0.02083928, 0.0159863 , 0.1464459 , 0.04795889, 0.06622895, 0.0262632 , 0.12988867, 0. , 0.09192121, 0.03111619, 0.01712818, 0.04852983, 0.05937768, 0.07079646, 0.04396232, 0.0225521 , 0.05994862, 0.01855552]])

-
calc_H(scale='eisenberg')[source]¶ Method for calculating global hydrophobicity (Eisenberg scale) of all sequences in the library.
- Parameters
scale – {str} hydrophobicity scale to use. For available scales, see
modlamp.descriptors.PeptideDescriptor.- Returns
{numpy.ndarray} Eisenberg hydrophobicities in the attribute
H.
-
calc_uH(window=1000, angle=100, modality='max')[source]¶ Method for calculating hydrophobic moments (Eisenberg scale) for all sequences in the library.
- Parameters
window – {int} amino acid window in which to calculate the moment. If the sequence is shorter than the window, the length of the sequence is taken. So if the default window of 1000 is chosen, for all sequences shorter than 1000, the global hydrophobic moment will be calculated. Otherwise, the maximal hydrophiobic moment for the chosen window size found in the sequence will be returned.
angle – {int} angle in which to calculate the moment. 100 for alpha helices, 180 for beta sheets.
modality – {‘max’ or ‘mean’} calculate respectively maximum or mean hydrophobic moment.
- Returns
{numpy.ndarray} calculated hydrophobic moments in the attribute
uH.
-
calc_charge(ph=7.0, amide=True)[source]¶ Method to calculate the total molecular charge at a given pH for all sequences in the library.
- Parameters
ph – {float} ph at which to calculate the peptide charge.
amide – {boolean} whether the sequences have an amidated C-terminus (-> charge += 1).
- Returns
{numpy.ndarray} calculated charges in the attribute
charge.
-
calc_len()[source]¶ Method to get the sequence length of all sequences in the library.
- Returns
{numpy.ndarray} sequence lengths in the attribute
len.
-
plot_summary(filename=None, colors=None, plot=True)[source]¶ Method to generate a visual summary of different characteristics of the given library. The class methods are used with their standard options.
- Parameters
filename – {str} path to save the generated plot to.
colors – {str / list} color or list of colors to use for plotting. e.g. ‘#4E395D’, ‘red’, ‘k’
plot – {boolean} whether the plot should be created or just the features are calculated
- Returns
visual summary (plot) of the library characteristics (if
plot=True).- Example
>>> g = GlobalAnalysis([seqs1, seqs2, seqs3]) # seqs being lists / arrays of sequences >>> g.plot_summary()
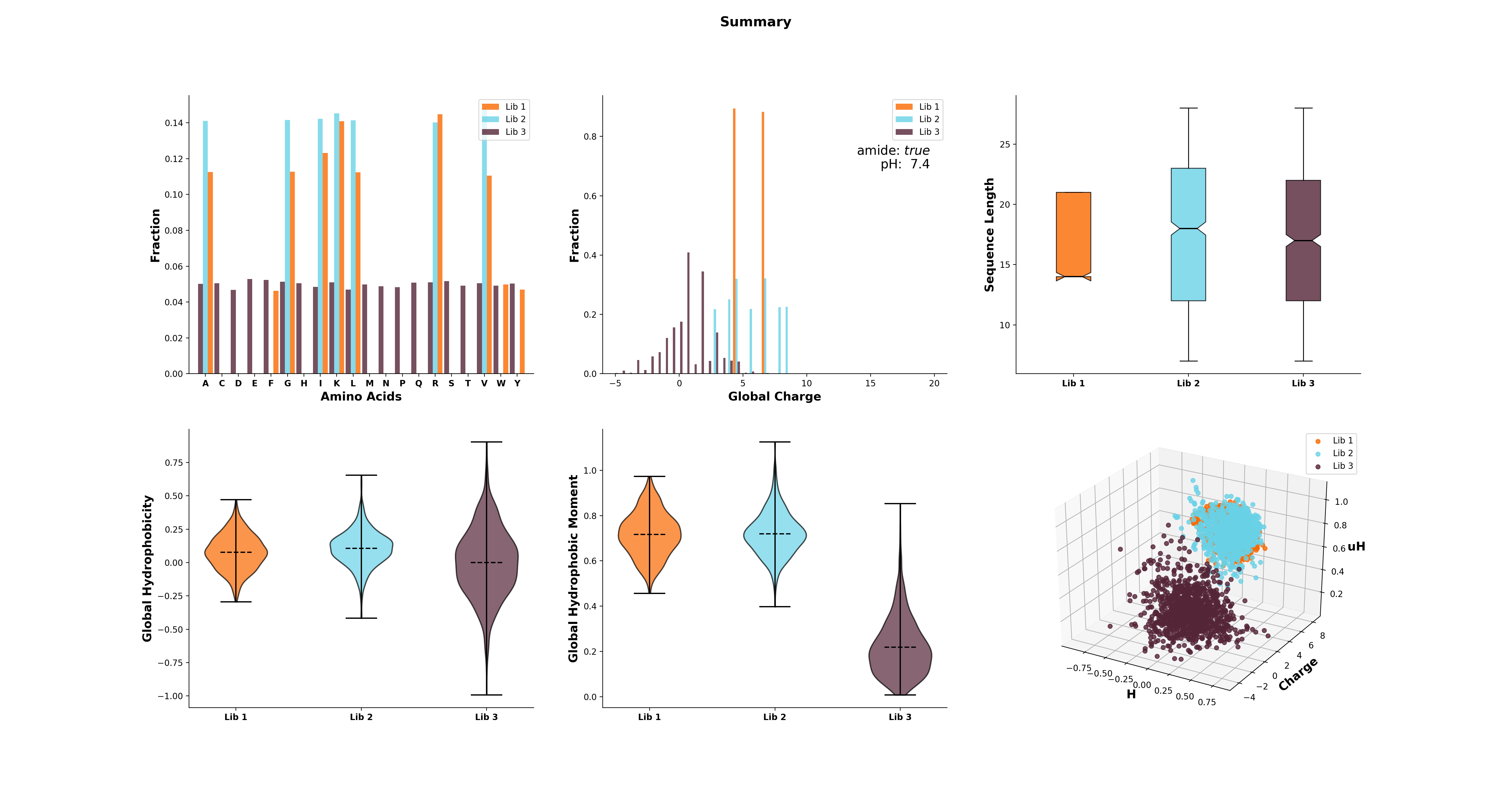
-
2.9. modlamp.core¶
Core helper functions and classes for other modules. The two main classes are:
Class |
Characteristics |
|---|---|
Base class inheriting to all sequence classes in the module |
|
Base class inheriting to the two descriptor classes in |
-
class
modlamp.core.BaseSequence(seqnum, lenmin=7, lenmax=28)[source]¶ Base class for sequence classes in the module
modlamp.sequences. It contains amino acid probabilities for different sequence generation classes.The following amino acid probabilities are used: (extracted from the APD3, March 17, 2016)
AA
rand
AMP
AMPnoCM
randnoCM
A
0.05
0.0766
0.0812275
0.05555555
C
0.05
0.071
0.0
0.0
D
0.05
0.026
0.0306275
0.05555555
E
0.05
0.0264
0.0310275
0.05555555
F
0.05
0.0405
0.0451275
0.05555555
G
0.05
0.1172
0.1218275
0.05555555
H
0.05
0.021
0.0256275
0.05555555
I
0.05
0.061
0.0656275
0.05555555
K
0.05
0.0958
0.1004275
0.05555555
L
0.05
0.0838
0.0884275
0.05555555
M
0.05
0.0123
0.0
0.0
N
0.05
0.0386
0.0432275
0.05555555
P
0.05
0.0463
0.0509275
0.05555555
Q
0.05
0.0251
0.0297275
0.05555555
R
0.05
0.0545
0.0591275
0.05555555
S
0.05
0.0613
0.0659275
0.05555555
T
0.05
0.0455
0.0501275
0.05555555
V
0.05
0.0572
0.0618275
0.05555555
W
0.05
0.0155
0.0201275
0.05555555
Y
0.05
0.0244
0.0290275
0.05555555
-
__init__(seqnum, lenmin=7, lenmax=28)[source]¶ - Parameters
seqnum – number of sequences to generate
lenmin – minimal length of the generated sequences
lenmax – maximal length of the generated sequences
- Returns
attributes
seqnum,lenminandlenmax.- Example
>>> b = BaseSequence(10, 7, 28) >>> b.seqnum 10 >>> b.lenmin 7 >>> b.lenmax 28
-
save_fasta(filename, names=False)[source]¶ Method to save generated sequences in a
.FASTAformatted file.- Parameters
filename – output filename in which the sequences from
sequencesare safed in fasta format.names – {bool} whether sequence names from
namesshould be saved as sequence identifiers
- Returns
a FASTA formatted file containing the generated sequences
- Example
>>> b = BaseSequence(2) >>> b.sequences = ['KLLSLSLALDLLS', 'KLPERTVVNSSDF'] >>> b.names = ['Sequence1', 'Sequence2'] >>> b.save_fasta('/location/of/fasta/file.fasta', names=True)
-
mutate_AA(nr, prob)[source]¶ Method to mutate with prob probability a nr of positions per sequence randomly.
- Parameters
nr – number of mutations to perform per sequence
prob – probability of mutating a sequence
- Returns
mutated sequences in the attribute
sequences.- Example
>>> b = BaseSequence(1) >>> b.sequences = ['IAKAGRAIIK'] >>> b.mutate_AA(3, 1.) >>> b.sequences ['NAKAGRAWIK']
-
filter_duplicates()[source]¶ Method to filter duplicates in the sequences from the class attribute
sequences- Returns
filtered sequences list in the attribute
sequencesand corresponding names.- Example
>>> b = BaseSequence(4) >>> b.sequences = ['KLLKLLKKLLKLLK', 'KLLKLLKKLLKLLK', 'KLAKLAKKLAKLAK', 'KLAKLAKKLAKLAK'] >>> b.filter_duplicates() >>> b.sequences ['KLLKLLKKLLKLLK', 'KLAKLAKKLAKLAK']
New in version v2.2.5.
-
keep_natural_aa()[source]¶ Method to filter out sequences that do not contain natural amino acids. If the sequence contains a character that is not in
['A','C','D,'E','F','G','H','I','K','L','M','N','P','Q','R','S','T','V','W','Y'].- Returns
filtered sequence list in the attribute
sequences. The other attributes are also filtered accordingly (if present).- Example
>>> b = BaseSequence(2) >>> b.sequences = ['BBBsdflUasUJfBJ', 'GLFDIVKKVVGALGSL'] >>> b.keep_natural_aa() >>> b.sequences ['GLFDIVKKVVGALGSL']
-
filter_aa(amino_acids)[source]¶ Method to filter out corresponding names and descriptor values of sequences with given amino acids in the argument list aminoacids.
- Parameters
amino_acids – {list} amino acids to be filtered
- Returns
filtered list of sequences names in the corresponding attributes.
- Example
>>> b = BaseSequence(3) >>> b.sequences = ['AAALLLIIIKKK', 'CCEERRT', 'LLVVIIFFFQQ'] >>> b.filter_aa(['C']) >>> b.sequences ['AAALLLIIIKKK', 'LLVVIIFFFQQ']
-
-
class
modlamp.core.BaseDescriptor(seqs)[source]¶ Base class inheriting to both peptide descriptor classes
modlamp.descriptors.GlobalDescriptorandmodlamp.descriptors.PeptideDescriptor.-
__init__(seqs)[source]¶ - Parameters
seqs – a
.FASTAfile with sequences, a list / array of sequences or a single sequence as string to calculate the descriptor values for.- Returns
initialized attributes
sequencesandnames.- Example
>>> AMP = BaseDescriptor('KLLKLLKKLLKLLK','pepCATS') >>> AMP.sequences ['KLLKLLKKLLKLLK'] >>> seqs = BaseDescriptor('/Path/to/file.fasta', 'eisenberg') # load sequences from .fasta file >>> seqs.sequences ['AFDGHLKI','KKLQRSDLLRTK','KKLASCNNIPPR'...]
-
read_fasta(filename)[source]¶ Method for loading sequences from a
.FASTAformatted file into the attributessequencesandnames.- Parameters
filename – {str}
.FASTAfile with sequences and headers to read- Returns
{list} sequences in the attribute
sequenceswith corresponding sequence names innames.
-
save_fasta(filename, names=False)[source]¶ Method for saving sequences from
sequencesto a.FASTAformatted file.- Parameters
filename – {str} filename of the output
.FASTAfilenames – {bool} whether sequence names from self.names should be saved as sequence identifiers
- Returns
a FASTA formatted file containing the generated sequences
-
count_aa(scale='relative', average=False, append=False)[source]¶ Method for producing the amino acid distribution for the given sequences as a descriptor
- Parameters
scale – {‘absolute’ or ‘relative’} defines whether counts or frequencies are given for each AA
average – {boolean} whether the averaged amino acid counts for all sequences should be returned
append – {boolean} whether the produced descriptor values should be appended to the existing ones in the attribute
descriptor.
- Returns
the amino acid distributions for every sequence individually in the attribute
descriptor- Example
>>> AMP = PeptideDescriptor('ACDEFGHIKLMNPQRSTVWY') # aa_count() does not depend on the descriptor scale >>> AMP.count_aa() >>> AMP.descriptor array([[ 0.05, 0.05, 0.05, 0.05, 0.05, 0.05, 0.05, 0.05, 0.05, ... ]]) >>> AMP.descriptor.shape (1, 20)
See also
modlamp.core.count_aa()
-
count_ngrams(n)[source]¶ Method for producing n-grams of all sequences in self.sequences
- Parameters
n – {int or list of ints} defines whether counts or frequencies are given for each AA
- Returns
{dict} dictionary with n-grams as keys and their counts in the sequence as values in
descriptor- Example
>>> D = PeptideDescriptor('GLLDFLSLAALSLDKLVKKGALS') >>> D.count_ngrams([2, 3]) >>> D.descriptor {'LS': 3, 'LD': 2, 'LSL': 2, 'AL': 2, ..., 'LVK': 1}
See also
-
feature_scaling(stype='standard', fit=True)[source]¶ Method for feature scaling of the calculated descriptor matrix.
- Parameters
stype – {‘standard’ or ‘minmax’} type of scaling to be used
fit – {boolean} defines whether the used scaler is first fitting on the data (True) or whether the already fitted scaler in
scalershould be used to transform (False).
- Returns
scaled descriptor values in
descriptor- Example
>>> D.descriptor array([[0.155],[0.34],[0.16235294],[-0.08842105],[0.116]]) >>> D.feature_scaling(type='minmax',fit=True) array([[0.56818182],[1.],[0.5853447],[0.],[0.47714988]])
-
feature_shuffle()[source]¶ Method for shuffling feature columns randomly.
- Returns
descriptor matrix with shuffled feature columns in
descriptor- Example
>>> D.descriptor array([[0.80685625,167.05234375,39.56818125,-0.26338667,155.16888667,33.48778]]) >>> D.feature_shuffle() array([[155.16888667,-0.26338667,167.05234375,0.80685625,39.56818125,33.48778]])
-
sequence_order_shuffle()[source]¶ Method for shuffling sequence order in the attribute
sequences.- Returns
sequences in
sequenceswith shuffled order in the list.- Example
>>> D.sequences ['LILRALKGAARALKVA','VKIAKIALKIIKGLG','VGVRLIKGIGRVARGAI','LRGLRGVIRGGKAIVRVGK','GGKLVRLIARIGKGV'] >>> D.sequence_order_shuffle() >>> D.sequences ['VGVRLIKGIGRVARGAI','LILRALKGAARALKVA','LRGLRGVIRGGKAIVRVGK','GGKLVRLIARIGKGV','VKIAKIALKIIKGLG']
-
random_selection(num)[source]¶ Method to randomly select a specified number of sequences (with names and descriptors if present) out of a given descriptor instance.
- Parameters
num – {int} number of entries to be randomly selected
- Returns
updated instance
- Example
>>> h = Helices(7, 28, 100) >>> h.generate_helices() >>> desc = PeptideDescriptor(h.sequences, 'eisenberg') >>> desc.calculate_moment() >>> len(desc.sequences) 100 >>> len(desc.descriptor) 100 >>> desc.random_selection(10) >>> len(desc.descriptor) 10 >>> len(desc.descriptor) 10
New in version v2.2.3.
-
minmax_selection(iterations, distmetric='euclidean', seed=0)[source]¶ Method to select a specified number of sequences according to the minmax algorithm.
- Parameters
iterations – {int} Number of sequences to retrieve.
distmetric – Distance metric to calculate the distances between the sequences in descriptor space. Choose from ‘euclidean’ or ‘minkowsky’.
seed – {int} Set a random seed for numpy to pick the first sequence.
- Returns
updated instance
-
filter_sequences(sequences)[source]¶ Method to filter out entries for given sequences in sequences out of a descriptor instance. All corresponding attribute values of these sequences (e.g. in
descriptor,name) are deleted as well. The method returns an updated descriptor instance.- Parameters
sequences – {list} sequences to be filtered out of the whole instance, including corresponding data
- Returns
updated instance without filtered sequences
- Example
>>> sequences = ['KLLKLLKKLLKLLK', 'ACDEFGHIK', 'GLFDIVKKVV', 'GLFDIVKKVVGALG', 'GLFDIVKKVVGALGSL'] >>> desc = PeptideDescriptor(sequences, 'pepcats') >>> desc.calculate_crosscorr(7) >>> len(desc.descriptor) 5 >>> desc.filter_sequences('KLLKLLKKLLKLLK') >>> len(desc.descriptor) 4 >>> desc.sequences ['ACDEFGHIK', 'GLFDIVKKVV', 'GLFDIVKKVVGALG', 'GLFDIVKKVVGALGSL']
-
filter_values(values, operator='==')[source]¶ Method to filter the descriptor matrix in the attribute
descriptorfor a given list of values (same size as the number of features in the descriptor matrix!) The operator option tells the method whether to filter for values equal, lower, higher ect. to the given values in the values array.- Parameters
values – {list} values to filter the attribute
descriptorforoperator – {str} filter criterion, available the operators
==,<,>,<=``and ``>=.
- Returns
descriptor matrix and updated sequences containing only entries with descriptor values given in values in the corresponding attributes.
- Example
>>> desc.descriptor # desc = BaseDescriptor instance array([[ 0.7666517 ], [ 0.38373498]]) >>> desc.filter_values([0.5], '<') >>> desc.descriptor array([[ 0.38373498]])
-
filter_aa(amino_acids)[source]¶ Method to filter out corresponding names and descriptor values of sequences with given amino acids in the argument list aminoacids.
- Parameters
amino_acids – list of amino acids to be filtered
- Returns
filtered list of sequences, descriptor values, target values and names in the corresponding attributes.
- Example
>>> b = BaseSequence(3) >>> b.sequences = ['AAALLLIIIKKK', 'CCEERRT', 'LLVVIIFFFQQ'] >>> b.filter_aa(['C']) >>> b.sequences ['AAALLLIIIKKK', 'LLVVIIFFFQQ']
-
filter_duplicates()[source]¶ Method to filter duplicates in the sequences from the class attribute
sequences- Returns
filtered sequences list in the attribute
sequencesand corresponding names.- Example
>>> b = BaseDescriptor(['KLLKLLKKLLKLLK', 'KLLKLLKKLLKLLK', 'KLAKLAKKLAKLAK', 'KLAKLAKKLAKLAK']) >>> b.filter_duplicates() >>> b.sequences ['KLLKLLKKLLKLLK', 'KLAKLAKKLAKLAK']
New in version v2.2.5.
-
keep_natural_aa()[source]¶ Method to filter out sequences that do not contain natural amino acids. If the sequence contains a character that is not in [‘A’,’C’,’D,’E’,’F’,’G’,’H’,’I’,’K’,’L’,’M’,’N’,’P’,’Q’,’R’,’S’,’T’,’V’,’W’,’Y’].
- Returns
filtered sequence list in the attribute
sequences. The other attributes are also filtered accordingly (if present).- Example
>>> b = BaseSequence(2) >>> b.sequences = ['BBBsdflUasUJfBJ', 'GLFDIVKKVVGALGSL'] >>> b.keep_natural_aa() >>> b.sequences ['GLFDIVKKVVGALGSL']
-
load_descriptordata(filename, delimiter=', ', targets=False, skip_header=0)[source]¶ Method to load any data file with sequences and descriptor values and save it to a new insatnce of the class
modlamp.descriptors.PeptideDescriptor.Note
Headers are not considered. To skip initial lines in the file, use the skip_header option.
- Parameters
filename – {str} filename of the data file to be loaded
delimiter – {str} column delimiter
targets – {boolean} whether last column in the file contains a target class vector
skip_header – {int} number of initial lines to skip in the file
- Returns
loaded sequences, descriptor values and targets in the corresponding attributes.
-
save_descriptor(filename, delimiter=', ', targets=None, header=None)[source]¶ Method to save the descriptor values to a .csv/.txt file
- Parameters
filename – filename of the output file
delimiter – column delimiter
targets – target class vector to be added to descriptor (same length as
sequences)header – {str} header to be written at the beginning of the file (if
None: feature names are taken)
- Returns
output file with peptide names and descriptor values
-
-
modlamp.core.load_scale(scalename)[source]¶ Method to load scale values for a given amino acid scale
- Parameters
scalename – amino acid scale name, for available scales see the
modlamp.descriptors.PeptideDescriptor()documentation.- Returns
amino acid scale values in dictionary format.
-
modlamp.core.read_fasta(inputfile)[source]¶ Method for loading sequences from a FASTA formatted file into
sequences&names. This method is used by the base classmodlamp.descriptors.PeptideDescriptorif the input is a FASTA file.- Parameters
inputfile – .fasta file with sequences and headers to read
- Returns
list of sequences in the attribute
sequenceswith corresponding sequence names innames.
-
modlamp.core.save_fasta(filename, sequences, names=None)[source]¶ Method for saving sequences in the instance
sequencesto a file in FASTA format.- Parameters
filename – {str} output filename (ending .fasta)
sequences – {list} sequences to be saved to file
names – {list} whether sequence names from self.names should be saved as sequence identifiers
- Returns
a FASTA formatted file containing the generated sequences
-
modlamp.core.aa_weights()[source]¶ Function holding molecular weight data on all natural amino acids.
- Returns
dictionary with amino acid letters and corresponding weights
New in version v2.4.1.
-
modlamp.core.count_aas(seq, scale='relative')[source]¶ Function to count the amino acids occuring in a given sequence.
- Parameters
seq – {str} amino acid sequence
scale – {‘absolute’ or ‘relative’} defines whether counts or frequencies are given for each AA
- Returns
{dict} dictionary with amino acids as keys and their counts in the sequence as values.
-
modlamp.core.count_ngrams(seq, n)[source]¶ Function to count the n-grams of an amino acid sequence. N can be one integer or a list of integers
- Parameters
seq – {str} amino acid sequence
n – {int or list of ints} defines whether counts or frequencies are given for each AA
- Returns
{dict} dictionary with n-grams as keys and their counts in the sequence as values.
-
modlamp.core.aa_energies()[source]¶ Function holding free energies of transfer between cyclohexane and water for all natural amino acids. H. G. Boman, D. Wade, I. a Boman, B. Wåhlin, R. B. Merrifield, FEBS Lett. 1989, 259, 103–106.
- Returns
dictionary with amino acid letters and corresponding energies.
-
modlamp.core.ngrams_apd()[source]¶ Function returning the most frequent 2-, 3- and 4-grams from all sequences in the APD3, version August 2016 with 2727 sequences. For all 2, 3 and 4grams, all possible ngrams were generated from all sequences and the top 50 most frequent assembled into a list. Finally, leading and tailing spaces were striped and duplicates as well as ngrams containing spaces were removed.
- Returns
numpy.array containing most frequent ngrams